Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses
- PMID: 24741084
- PMCID: PMC4054416
- DOI: 10.1128/JVI.00591-14
Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses
Abstract
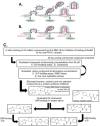
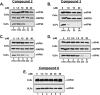
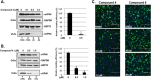
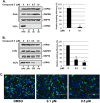
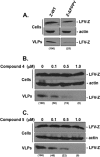
Budding of filoviruses, arenaviruses, and rhabdoviruses is facilitated by subversion of host proteins, such as Nedd4 E3 ubiquitin ligase, by viral PPxY late (L) budding domains expressed within the matrix proteins of these RNA viruses. As L domains are important for budding and are highly conserved in a wide array of RNA viruses, they represent potential broad-spectrum targets for the development of antiviral drugs. To identify potential competitive blockers, we used the known Nedd4 WW domain-PPxY interaction interface as the basis of an in silico screen. Using PPxY-dependent budding of Marburg (MARV) VP40 virus-like particles (VLPs) as our model system, we identified small-molecule hit 1 that inhibited Nedd4-PPxY interaction and PPxY-dependent budding. This lead candidate was subsequently improved with additional structure-activity relationship (SAR) analog testing which enhanced antibudding activity into the nanomolar range. Current lead compounds 4 and 5 exhibit on-target effects by specifically blocking the MARV VP40 PPxY-host Nedd4 interaction and subsequent PPxY-dependent egress of MARV VP40 VLPs. In addition, lead compounds 4 and 5 exhibited antibudding activity against Ebola and Lassa fever VLPs, as well as vesicular stomatitis and rabies viruses (VSV and RABV, respectively). These data provide target validation and suggest that inhibition of the PPxY-Nedd4 interaction can serve as the basis for the development of a novel class of broad-spectrum, host-oriented antivirals targeting viruses that depend on a functional PPxY L domain for efficient egress.
Importance: There is an urgent and unmet need for the development of safe and effective therapeutics against biodefense and high-priority pathogens, including filoviruses (Ebola and Marburg) and arenaviruses (e.g., Lassa and Junin) which cause severe hemorrhagic fever syndromes with high mortality rates. We along with others have established that efficient budding of filoviruses, arenaviruses, and other viruses is critically dependent on the subversion of host proteins. As disruption of virus budding would prevent virus dissemination, identification of small-molecule compounds that block these critical viral-host interactions should effectively block disease progression and transmission. Our findings provide validation for targeting these virus-host interactions as we have identified lead inhibitors with broad-spectrum antiviral activity. In addition, such inhibitors might prove useful for newly emerging RNA viruses for which no therapeutics would be available.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Chaperone-Mediated Autophagy Protein BAG3 Negatively Regulates Ebola and Marburg VP40-Mediated Egress.PLoS Pathog. 2017 Jan 11;13(1):e1006132. doi: 10.1371/journal.ppat.1006132. eCollection 2017 Jan. PLoS Pathog. 2017. PMID: 28076420 Free PMC article.
-
Ubiquitin Ligase WWP1 Interacts with Ebola Virus VP40 To Regulate Egress.J Virol. 2017 Sep 27;91(20):e00812-17. doi: 10.1128/JVI.00812-17. Print 2017 Oct 15. J Virol. 2017. PMID: 28768865 Free PMC article.
-
Angiomotin Counteracts the Negative Regulatory Effect of Host WWOX on Viral PPxY-Mediated Egress.J Virol. 2021 Mar 25;95(8):e00121-21. doi: 10.1128/JVI.00121-21. Epub 2021 Feb 3. J Virol. 2021. PMID: 33536174 Free PMC article.
-
Host and Viral Proteins Modulating Ebola and Marburg Virus Egress.Viruses. 2019 Jan 3;11(1):25. doi: 10.3390/v11010025. Viruses. 2019. PMID: 30609802 Free PMC article. Review.
-
Viruses go modular.J Biol Chem. 2020 Apr 3;295(14):4604-4616. doi: 10.1074/jbc.REV119.012414. Epub 2020 Feb 28. J Biol Chem. 2020. PMID: 32111739 Free PMC article. Review.
Cited by
-
Therapeutics for postexposure treatment of Ebola virus infection.Future Virol. 2015 Mar;10(3):221-232. doi: 10.2217/fvl.14.109. Future Virol. 2015. PMID: 26213559 Free PMC article.
-
ALIX Rescues Budding of a Double PTAP/PPEY L-Domain Deletion Mutant of Ebola VP40: A Role for ALIX in Ebola Virus Egress.J Infect Dis. 2015 Oct 1;212 Suppl 2(Suppl 2):S138-45. doi: 10.1093/infdis/jiu838. Epub 2015 Mar 18. J Infect Dis. 2015. PMID: 25786915 Free PMC article.
-
Covalent Tethering of Fragments For Covalent Probe Discovery.Medchemcomm. 2016 Apr 1;7(4):576-585. doi: 10.1039/c5md00518c. Epub 2016 Jan 28. Medchemcomm. 2016. PMID: 27398190 Free PMC article.
-
Influence of cellular trafficking pathway on bluetongue virus infection in ovine cells.Viruses. 2015 May 13;7(5):2378-403. doi: 10.3390/v7052378. Viruses. 2015. PMID: 25984713 Free PMC article.
-
Chaperone-Mediated Autophagy Protein BAG3 Negatively Regulates Ebola and Marburg VP40-Mediated Egress.PLoS Pathog. 2017 Jan 11;13(1):e1006132. doi: 10.1371/journal.ppat.1006132. eCollection 2017 Jan. PLoS Pathog. 2017. PMID: 28076420 Free PMC article.
References
-
- Feldmann H, Klenk HD. 1996. Filoviruses, p 877–888 In Baron S. (ed), Medical microbiology, 4th ed. University of Texas Medical Branch at Galveston, Galveston, TX - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

