Identification of a diguanylate cyclase and its role in Porphyromonas gingivalis virulence
- PMID: 24733094
- PMCID: PMC4097614
- DOI: 10.1128/IAI.00084-14
Identification of a diguanylate cyclase and its role in Porphyromonas gingivalis virulence
Abstract
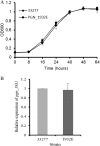
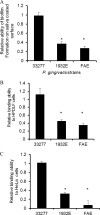
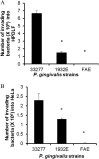
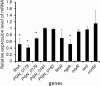
Porphyromonas gingivalis is a Gram-negative obligate anaerobic bacterium and is considered a keystone pathogen in the initiation of periodontitis, one of the most widespread infectious diseases. Bacterial bis-(3'-5') cyclic GMP (cyclic di-GMP [c-di-GMP]) serves as a second messenger and is involved in modulating virulence factors in numerous bacteria. However, the role of this second messenger has not been investigated in P. gingivalis, mainly due to a lack of an annotation regarding diguanylate cyclases (DGCs) in this bacterium. Using bioinformatics tools, we found a protein, PGN_1932, containing a GGDEF domain. A deletion mutation in the pgn_1932 gene had a significant effect on the intracellular c-di-GMP level in P. gingivalis. Genetic analysis showed that expression of the fimA and rgpA genes, encoding the major protein subunit of fimbriae and an arginine-specific proteinase, respectively, was downregulated in the pgn_1932 mutant. Correspondingly, FimA protein production and the fimbrial display on the mutant were significantly reduced. Mutations in the pgn_1932 gene also had a significant impact on the adhesive and invasive capabilities of P. gingivalis, which are required for its pathogenicity. These findings provide evidence that the PGN_1932 protein is both responsible for synthesizing c-di-GMP and involved in biofilm formation and host cell invasion by P. gingivalis by controlling the expression and biosynthesis of FimA.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Altered Regulation of the Diguanylate Cyclase YaiC Reduces Production of Type 1 Fimbriae in a Pst Mutant of Uropathogenic Escherichia coli CFT073.J Bacteriol. 2017 Nov 14;199(24):e00168-17. doi: 10.1128/JB.00168-17. Print 2017 Dec 15. J Bacteriol. 2017. PMID: 28924030 Free PMC article.
-
Thermoregulation of Biofilm Formation in Burkholderia pseudomallei Is Disrupted by Mutation of a Putative Diguanylate Cyclase.J Bacteriol. 2017 Feb 14;199(5):e00780-16. doi: 10.1128/JB.00780-16. Print 2017 Mar 1. J Bacteriol. 2017. PMID: 27956524 Free PMC article.
-
Global Transcriptional Repression of Diguanylate Cyclases by MucR1 Is Essential for Sinorhizobium-Soybean Symbiosis.mBio. 2021 Oct 26;12(5):e0119221. doi: 10.1128/mBio.01192-21. Epub 2021 Oct 26. mBio. 2021. PMID: 34700374 Free PMC article.
-
Diguanylate Cyclases in Vibrio cholerae: Essential Regulators of Lifestyle Switching.Front Cell Infect Microbiol. 2020 Oct 22;10:582947. doi: 10.3389/fcimb.2020.582947. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33194821 Free PMC article. Review.
-
Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease.J Periodontol. 2003 Jan;74(1):90-6. doi: 10.1902/jop.2003.74.1.90. J Periodontol. 2003. PMID: 12593602 Review.
Cited by
-
The Oxidative Stress-Induced Hypothetical Protein PG_0686 in Porphyromonas gingivalis W83 Is a Novel Diguanylate Cyclase.Microbiol Spectr. 2023 Jan 31;11(2):e0441122. doi: 10.1128/spectrum.04411-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36719196 Free PMC article.
-
Cyclic Dinucleotides in Oral Bacteria and in Oral Biofilms.Front Cell Infect Microbiol. 2017 Jun 21;7:273. doi: 10.3389/fcimb.2017.00273. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28680857 Free PMC article.
-
[Cloning, expression, and purification of c-di-AMP metabolism-related genes from Porphyromonas gingivalis].Hua Xi Kou Qiang Yi Xue Za Zhi. 2015 Dec;33(6):607-12. doi: 10.7518/hxkq.2015.06.012. Hua Xi Kou Qiang Yi Xue Za Zhi. 2015. PMID: 27051954 Free PMC article. Chinese.
-
Fimbriae-mediated outer membrane vesicle production and invasion of Porphyromonas gingivalis.Microbiologyopen. 2015 Feb;4(1):53-65. doi: 10.1002/mbo3.221. Epub 2014 Dec 18. Microbiologyopen. 2015. PMID: 25524808 Free PMC article.
-
Atypical cyclic di-AMP signaling is essential for Porphyromonas gingivalis growth and regulation of cell envelope homeostasis and virulence.NPJ Biofilms Microbiomes. 2022 Jul 6;8(1):53. doi: 10.1038/s41522-022-00316-w. NPJ Biofilms Microbiomes. 2022. PMID: 35794154 Free PMC article.
References
-
- Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416–4425 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

