First comprehensive in silico analysis of the functional and structural consequences of SNPs in human GalNAc-T1 gene
- PMID: 24723968
- PMCID: PMC3960557
- DOI: 10.1155/2014/904052
First comprehensive in silico analysis of the functional and structural consequences of SNPs in human GalNAc-T1 gene
Abstract
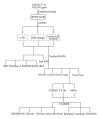
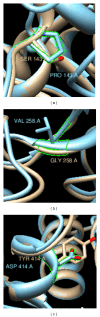
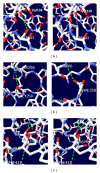
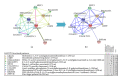
GalNAc-T1, a key candidate of GalNac-transferases genes family that is involved in mucin-type O-linked glycosylation pathway, is expressed in most biological tissues and cell types. Despite the reported association of GalNAc-T1 gene mutations with human disease susceptibility, the comprehensive computational analysis of coding, noncoding and regulatory SNPs, and their functional impacts on protein level, still remains unknown. Therefore, sequence- and structure-based computational tools were employed to screen the entire listed coding SNPs of GalNAc-T1 gene in order to identify and characterize them. Our concordant in silico analysis by SIFT, PolyPhen-2, PANTHER-cSNP, and SNPeffect tools, identified the potential nsSNPs (S143P, G258V, and Y414D variants) from 18 nsSNPs of GalNAc-T1. Additionally, 2 regulatory SNPs (rs72964406 and #x26; rs34304568) were also identified in GalNAc-T1 by using FastSNP tool. Using multiple computational approaches, we have systematically classified the functional mutations in regulatory and coding regions that can modify expression and function of GalNAc-T1 enzyme. These genetic variants can further assist in better understanding the wide range of disease susceptibility associated with the mucin-based cell signalling and pathogenic binding, and may help to develop novel therapeutic elements for associated diseases.
Figures



Similar articles
-
Impacts of Nonsynonymous Single Nucleotide Polymorphisms of Adiponectin Receptor 1 Gene on Corresponding Protein Stability: A Computational Approach.Biomed Res Int. 2016;2016:9142190. doi: 10.1155/2016/9142190. Epub 2016 May 15. Biomed Res Int. 2016. PMID: 27294143 Free PMC article.
-
In-Silico Computing of the Most Deleterious nsSNPs in HBA1 Gene.PLoS One. 2016 Jan 29;11(1):e0147702. doi: 10.1371/journal.pone.0147702. eCollection 2016. PLoS One. 2016. PMID: 26824843 Free PMC article.
-
Identification of functional SNPs in BARD1 gene and in silico analysis of damaging SNPs: based on data procured from dbSNP database.PLoS One. 2012;7(10):e43939. doi: 10.1371/journal.pone.0043939. Epub 2012 Oct 9. PLoS One. 2012. PMID: 23056176 Free PMC article.
-
Lost in the space of bioinformatic tools: a constantly updated survival guide for genetic epidemiology. The GenEpi Toolbox.Atherosclerosis. 2010 Apr;209(2):321-35. doi: 10.1016/j.atherosclerosis.2009.10.026. Epub 2009 Oct 29. Atherosclerosis. 2010. PMID: 19963217 Review.
-
Molecular genetic epidemiology of human diseases: from patterns to predictions.Hum Genet. 2014 Apr;133(4):425-30. doi: 10.1007/s00439-013-1396-y. Epub 2013 Nov 19. Hum Genet. 2014. PMID: 24241280 Review.
Cited by
-
Systematic exploration of predicted destabilizing nonsynonymous single nucleotide polymorphisms (nsSNPs) of human aldehyde oxidase: A Bio-informatics study.Pharmacol Res Perspect. 2019 Nov 22;7(6):e00538. doi: 10.1002/prp2.538. eCollection 2019 Dec. Pharmacol Res Perspect. 2019. PMID: 31768259 Free PMC article.
-
Identification genetic variations in some heat shock protein genes of Tali goat breed and study their structural and functional effects on relevant proteins.Vet Med Sci. 2023 Sep;9(5):2247-2259. doi: 10.1002/vms3.1231. Epub 2023 Aug 2. Vet Med Sci. 2023. PMID: 37530404 Free PMC article.
-
In silico prediction of deleterious non-synonymous SNPs in STAT3.Asian Biomed (Res Rev News). 2023 Oct 18;17(4):185-199. doi: 10.2478/abm-2023-0059. eCollection 2023 Aug. Asian Biomed (Res Rev News). 2023. PMID: 37860678 Free PMC article.
-
In silico analysis of consequences of non-synonymous SNPs of Slc11a2 gene in Indian bovines.Genom Data. 2015 May 30;5:72-9. doi: 10.1016/j.gdata.2015.05.015. eCollection 2015 Sep. Genom Data. 2015. PMID: 26484229 Free PMC article.
-
Computational Insights into the Deleterious Impacts of Missense Variants on N-Acetyl-d-glucosamine Kinase Structure and Function.Int J Mol Sci. 2021 Jul 28;22(15):8048. doi: 10.3390/ijms22158048. Int J Mol Sci. 2021. PMID: 34360815 Free PMC article.
References
-
- Bennett EP, Weghuis DO, Merkx G, van Kessel AG, Eiberg H, Clausen H. Genomic organization and chromosomal localization of three members of the UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferase family. Glycobiology. 1998;8(6):547–555. - PubMed
-
- Hang HC, Bertozzi CR. The chemistry and biology of mucin-type O-linked glycosylation. Bioorganic and Medicinal Chemistry. 2005;13(17):5021–5034. - PubMed
-
- Hussain MRM. The role of Galactose in human health and disease. Central European Journal of Medicine. 2012;7(4):409–419.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

