Copper is required for oncogenic BRAF signalling and tumorigenesis
- PMID: 24717435
- PMCID: PMC4138975
- DOI: 10.1038/nature13180
Copper is required for oncogenic BRAF signalling and tumorigenesis
Abstract
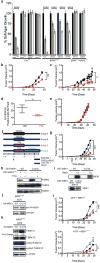
The BRAF kinase is mutated, typically Val 600→Glu (V600E), to induce an active oncogenic state in a large fraction of melanomas, thyroid cancers, hairy cell leukaemias and, to a smaller extent, a wide spectrum of other cancers. BRAF(V600E) phosphorylates and activates the MEK1 and MEK2 kinases, which in turn phosphorylate and activate the ERK1 and ERK2 kinases, stimulating the mitogen-activated protein kinase (MAPK) pathway to promote cancer. Targeting MEK1/2 is proving to be an important therapeutic strategy, given that a MEK1/2 inhibitor provides a survival advantage in metastatic melanoma, an effect that is increased when administered together with a BRAF(V600E) inhibitor. We previously found that copper (Cu) influx enhances MEK1 phosphorylation of ERK1/2 through a Cu-MEK1 interaction. Here we show decreasing the levels of CTR1 (Cu transporter 1), or mutations in MEK1 that disrupt Cu binding, decreased BRAF(V600E)-driven signalling and tumorigenesis in mice and human cell settings. Conversely, a MEK1-MEK5 chimaera that phosphorylated ERK1/2 independently of Cu or an active ERK2 restored the tumour growth of murine cells lacking Ctr1. Cu chelators used in the treatment of Wilson disease decreased tumour growth of human or murine cells transformed by BRAF(V600E) or engineered to be resistant to BRAF inhibition. Taken together, these results suggest that Cu-chelation therapy could be repurposed to treat cancers containing the BRAF(V600E) mutation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
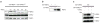

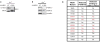

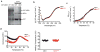


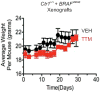
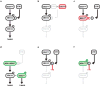

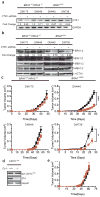
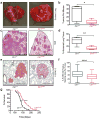
Comment in
-
Signalling: Inhibiting oncogenic BRAF signalling by copper depletion.Nat Rev Cancer. 2014 Jun;14(6):384-5. doi: 10.1038/nrc3745. Epub 2014 Apr 28. Nat Rev Cancer. 2014. PMID: 24769757 No abstract available.
-
Ctr1-ing BRAF signaling with copper.Pigment Cell Melanoma Res. 2014 Sep;27(5):689-91. doi: 10.1111/pcmr.12265. Epub 2014 Jun 6. Pigment Cell Melanoma Res. 2014. PMID: 24835481 No abstract available.
Similar articles
-
Inhibition of BCL2 Family Members Increases the Efficacy of Copper Chelation in BRAFV600E-Driven Melanoma.Cancer Res. 2020 Apr 1;80(7):1387-1400. doi: 10.1158/0008-5472.CAN-19-1784. Epub 2020 Jan 31. Cancer Res. 2020. PMID: 32005716 Free PMC article.
-
Copper Chelation as Targeted Therapy in a Mouse Model of Oncogenic BRAF-Driven Papillary Thyroid Cancer.Clin Cancer Res. 2018 Sep 1;24(17):4271-4281. doi: 10.1158/1078-0432.CCR-17-3705. Epub 2018 Jul 31. Clin Cancer Res. 2018. PMID: 30065097 Free PMC article.
-
Copper Chelation Inhibits BRAFV600E-Driven Melanomagenesis and Counters Resistance to BRAFV600E and MEK1/2 Inhibitors.Cancer Res. 2017 Nov 15;77(22):6240-6252. doi: 10.1158/0008-5472.CAN-16-1190. Epub 2017 Oct 6. Cancer Res. 2017. PMID: 28986383 Free PMC article.
-
That which does not kill me makes me stronger; combining ERK1/2 pathway inhibitors and BH3 mimetics to kill tumour cells and prevent acquired resistance.Br J Pharmacol. 2013 Aug;169(8):1708-22. doi: 10.1111/bph.12220. Br J Pharmacol. 2013. PMID: 23647573 Free PMC article. Review.
-
Targeting oncogenic Raf protein-serine/threonine kinases in human cancers.Pharmacol Res. 2018 Sep;135:239-258. doi: 10.1016/j.phrs.2018.08.013. Epub 2018 Aug 15. Pharmacol Res. 2018. PMID: 30118796 Review.
Cited by
-
Diagnostic and Dosimetry Features of [64Cu]CuCl2 in High-Grade Paediatric Infiltrative Gliomas.Mol Imaging Biol. 2023 Apr;25(2):391-400. doi: 10.1007/s11307-022-01769-3. Epub 2022 Aug 30. Mol Imaging Biol. 2023. PMID: 36042116
-
Recognition- and reactivity-based fluorescent probes for studying transition metal signaling in living systems.Acc Chem Res. 2015 Aug 18;48(8):2434-42. doi: 10.1021/acs.accounts.5b00221. Epub 2015 Jul 28. Acc Chem Res. 2015. PMID: 26215055 Free PMC article.
-
Comprehensive analysis of cuproptosis-related genes and tumor microenvironment infiltration characterization in breast cancer.Front Immunol. 2022 Oct 20;13:978909. doi: 10.3389/fimmu.2022.978909. eCollection 2022. Front Immunol. 2022. PMID: 36341328 Free PMC article.
-
Copper Homeostasis Based on Cuproptosis-Related Signature Optimizes Molecular Subtyping and Treatment of Glioma.Mol Neurobiol. 2024 Aug;61(8):4962-4975. doi: 10.1007/s12035-023-03893-9. Epub 2023 Dec 28. Mol Neurobiol. 2024. PMID: 38151613
-
Copper metabolism as a unique vulnerability in cancer.Biochim Biophys Acta Mol Cell Res. 2021 Feb;1868(2):118893. doi: 10.1016/j.bbamcr.2020.118893. Epub 2020 Oct 20. Biochim Biophys Acta Mol Cell Res. 2021. PMID: 33091507 Free PMC article. Review.
References
-
- Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. - PubMed
-
- Wan PT, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. - PubMed
-
- Flaherty KT, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K01 CA178145/CA/NCI NIH HHS/United States
- T32 GM007184/GM/NIGMS NIH HHS/United States
- R21 CA172104/CA/NCI NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- R01 DK074192/DK/NIDDK NIH HHS/United States
- 092809/Z/10/Z/WT_/Wellcome Trust/United Kingdom
- CA178145/CA/NCI NIH HHS/United States
- CA094184/CA/NCI NIH HHS/United States
- P01 HL075443/HL/NHLBI NIH HHS/United States
- P30 CA014236/CA/NCI NIH HHS/United States
- DK074192/DK/NIDDK NIH HHS/United States
- T32 GM008570/GM/NIGMS NIH HHS/United States
- CA172104/CA/NCI NIH HHS/United States
- 092809/Wellcome Trust/United Kingdom
- R01 CA094184/CA/NCI NIH HHS/United States
- HL075443/HL/NHLBI NIH HHS/United States
- R01 CA089614/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

