Characterization of the miiuy croaker (Miichthys miiuy) transcriptome and development of immune-relevant genes and molecular markers
- PMID: 24714210
- PMCID: PMC3979730
- DOI: 10.1371/journal.pone.0094046
Characterization of the miiuy croaker (Miichthys miiuy) transcriptome and development of immune-relevant genes and molecular markers
Abstract
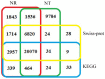
Background: The miiuy croaker (Miichthys miiuy) is an important species of marine fish that supports capture fisheries and aquaculture. At present commercial scale aquaculture of this species is limited due to diseases caused by pathogens and parasites which restrict production and limit commercial value. The lack of transcriptomic and genomic information for the miiuy croaker limits the ability of researchers to study the pathogenesis and immune system of this species. In this study we constructed a cDNA library from liver, spleen and kidney which was sequenced using Illumina paired-end sequencing to enable gene discovery and molecular marker development.
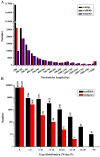
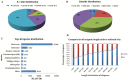
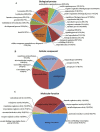
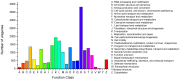
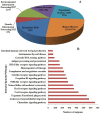
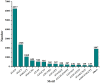
Principal findings: In our study, a total of 69,071 unigenes with an average length of 572 bp were obtained. Of these, 45,676 (66.13%) were successfully annotated in public databases. The unigenes were also annotated with Gene Ontology, Clusters of Orthologous Groups and KEGG pathways. Additionally, 498 immune-relevant genes were identified and classified. Furthermore, 14,885 putative simple sequence repeats (cSSRs) and 8,510 putative single nucleotide polymorphisms (SNPs) were identified from the 69,071 unigenes.
Conclusion: The miiuy croaker (Miichthys miiuy) transcriptome data provides a large resource to identify new genes involved in many processes including those involved in the response to pathogens and diseases. Furthermore, the thousands of potential cSSR and SNP markers found in this study are important resources with respect to future development of molecular marker assisted breeding programs for the miiuy croaker.
Conflict of interest statement
Figures
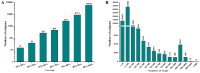

Similar articles
-
Genome-guided transcriptome analysis of miiuy croaker provides insights into pattern recognition receptors and cytokines in response to Vibrio anguillarum.Dev Comp Immunol. 2017 Aug;73:72-78. doi: 10.1016/j.dci.2017.03.009. Epub 2017 Mar 14. Dev Comp Immunol. 2017. PMID: 28315364
-
Identification of immune genes of the miiuy croaker (Miichthys miiuy) by sequencing and bioinformatic analysis of ESTs.Fish Shellfish Immunol. 2010 Dec;29(6):1099-105. doi: 10.1016/j.fsi.2010.08.013. Epub 2010 Aug 27. Fish Shellfish Immunol. 2010. PMID: 20801222
-
Comparative genomic of the teleost cathepsin B and H and involvement in bacterial induced immunity of miiuy croaker.Fish Shellfish Immunol. 2014 Dec;41(2):163-71. doi: 10.1016/j.fsi.2014.08.025. Epub 2014 Aug 27. Fish Shellfish Immunol. 2014. PMID: 25181651
-
Characterization, evolution and functional analysis of the liver-expressed antimicrobial peptide 2 (LEAP-2) gene in miiuy croaker.Fish Shellfish Immunol. 2014 Dec;41(2):191-9. doi: 10.1016/j.fsi.2014.08.010. Epub 2014 Aug 30. Fish Shellfish Immunol. 2014. PMID: 25180825
-
Sequence and expression analysis of cathepsin S gene in the miiuy croaker Miichthys miiuy.Fish Physiol Biochem. 2011 Dec;37(4):761-5. doi: 10.1007/s10695-011-9475-2. Epub 2011 Mar 20. Fish Physiol Biochem. 2011. PMID: 21424529
Cited by
-
Cytoprotective Effect of Antioxidant Pentapeptides from the Protein Hydrolysate of Swim Bladders of Miiuy Croaker (Miichthys miiuy) against H2O2-Mediated Human Umbilical Vein Endothelial Cell (HUVEC) Injury.Int J Mol Sci. 2019 Oct 31;20(21):5425. doi: 10.3390/ijms20215425. Int J Mol Sci. 2019. PMID: 31683554 Free PMC article.
-
Preparation, Physicochemical and Antioxidant Properties of Acid- and Pepsin-Soluble Collagens from the Swim Bladders of Miiuy Croaker (Miichthys miiuy).Mar Drugs. 2018 May 12;16(5):161. doi: 10.3390/md16050161. Mar Drugs. 2018. PMID: 29757239 Free PMC article.
-
Physicochemical and Antioxidant Properties of Acid- and Pepsin-Soluble Collagens from the Scales of Miiuy Croaker (Miichthys Miiuy).Mar Drugs. 2018 Oct 20;16(10):394. doi: 10.3390/md16100394. Mar Drugs. 2018. PMID: 30347803 Free PMC article.
-
Elucidating the Efficacy of Vaccination against Vibriosis in Lates calcarifer Using Two Recombinant Protein Vaccines Containing the Outer Membrane Protein K (r-OmpK) of Vibrio alginolyticus and the DNA Chaperone J (r-DnaJ) of Vibrio harveyi.Vaccines (Basel). 2020 Nov 6;8(4):660. doi: 10.3390/vaccines8040660. Vaccines (Basel). 2020. PMID: 33171991 Free PMC article.
-
SSREnricher: a computational approach for large-scale identification of polymorphic microsatellites based on comparative transcriptome analysis.PeerJ. 2020 Jul 2;8:e9372. doi: 10.7717/peerj.9372. eCollection 2020. PeerJ. 2020. PMID: 32676221 Free PMC article.
References
-
- Froese R, Pauly D (2012) FishBase:World Wide Web electronic publication. http://www.fishbase.org.
-
- Lou B (2004) Biology and breeding technology of Miichthys miiuy . Chin J Mod Fish 6: 11–13.
-
- Shan XJ, Dou SZ (2009) Allometric growth of croaker (Miichthys miiuy) larvae and juveniles and its ecological implication. Oceanologia et Limnologia Sinica 40: 714–718.
-
- Shan LZ, Xie QL, Shao XB, Yan MC (2010) Study on embryonic development and morphological characteristic habitual behavior for larvae, juvenile and young of Miichthys Miiuy . China Mar Sci 34: 75–80.
-
- Bondad-Reantaso MG (2007) Assessment of freshwater fish seed resources for sustainable aquaculture. Rome: Food and Agriculture Organization of the United Nations.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

