Nuclear import of SAMHD1 is mediated by a classical karyopherin α/β1 dependent pathway and confers sensitivity to VpxMAC induced ubiquitination and proteasomal degradation
- PMID: 24712655
- PMCID: PMC4098787
- DOI: 10.1186/1742-4690-11-29
Nuclear import of SAMHD1 is mediated by a classical karyopherin α/β1 dependent pathway and confers sensitivity to VpxMAC induced ubiquitination and proteasomal degradation
Abstract
Background: The deoxynucleotide-triphosphate (dNTP) hydrolase sterile alpha motif domain and HD domain 1 (SAMHD1) is a nuclear protein that inhibits HIV-1 infection in myeloid cells as well as quiescent CD4 T-cells, by decreasing the intracellular dNTP concentration below a level that is required for efficient reverse transcription. The Vpx proteins of the SIVSMM/HIV-2 lineage of lentiviruses bind SAMHD1 and recruit an ubiquitin ligase, leading to polyubiquitination and proteasomal degradation.
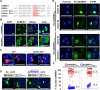
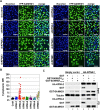
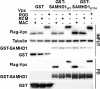
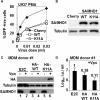
Results: Here, we have investigated the importance of nuclear localization for SAMHD1's antiviral function as well as its sensitivity to the Vpx protein of SIVMAC. Using GST pull down assays, as well as RNA silencing approaches, we show that SAMHD1 preferentially uses karyopherin α2 (KPNA2) and a classical N-terminal nuclear localization signal (14KRPR17) to enter the nucleus. Reduction of karyopherin β1 (KPNB1) or KPNA2 by RNAi also led to cytoplasmic re-distribution of SAMHD1. Using primary human monocyte-derived macrophages (MDM), a cell type in which SAMHD1 is naturally expressed to high levels, we demonstrate that nuclear localization is not required for its antiviral activity. Cytoplasmic SAMHD1 still binds to VpxMAC, is efficiently polyubiquitinated, but is not degraded. We also find that VpxMAC-induced SAMHD1 degradation was partially reversed by ubiquitin carrying the K48R or K11R substitution mutations, suggesting involvement of K48 and K11 linkages in SAMHD1 polyubiquitination. Using ubiquitin K-R mutants also revealed differences in the ubiquitin linkages between wild type and cytoplasmic forms of SAMHD1, suggesting a potential association with the resistance of cytoplasmic SAMHD1 to VpxMAC induced degradation.
Conclusions: Our work extends published observations on SAMHD1 nuclear localization to a natural cell type for HIV-1 infection, identifies KPNA2/KPNB1 as cellular proteins important for SAMHD1 nuclear import, and indicates that components of the nuclear proteasomal degradation machinery are required for SAMHD1 degradation.
Figures

Similar articles
-
Identification of critical regions in human SAMHD1 required for nuclear localization and Vpx-mediated degradation.PLoS One. 2013 Jul 11;8(7):e66201. doi: 10.1371/journal.pone.0066201. Print 2013. PLoS One. 2013. PMID: 23874389 Free PMC article.
-
The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus.J Virol. 2012 Dec;86(23):12552-60. doi: 10.1128/JVI.01657-12. Epub 2012 Sep 12. J Virol. 2012. PMID: 22973040 Free PMC article.
-
Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac.Retrovirology. 2012 Jun 12;9:49. doi: 10.1186/1742-4690-9-49. Retrovirology. 2012. PMID: 22691373 Free PMC article.
-
[Research Progress in Viral Protein Vpx induction of Proteasomal Degradation of the Antiviral Factor SAMHD1].Bing Du Xue Bao. 2016 May;32(3):355-60. Bing Du Xue Bao. 2016. PMID: 29963825 Review. Chinese.
-
Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase.Curr Opin Virol. 2012 Dec;2(6):755-63. doi: 10.1016/j.coviro.2012.09.010. Epub 2012 Oct 10. Curr Opin Virol. 2012. PMID: 23062609 Free PMC article. Review.
Cited by
-
The dNTPase activity of SAMHD1 is important for its suppression of innate immune responses in differentiated monocytic cells.J Biol Chem. 2020 Feb 7;295(6):1575-1586. doi: 10.1074/jbc.RA119.010360. Epub 2019 Dec 30. J Biol Chem. 2020. PMID: 31914403 Free PMC article.
-
The ability of SAMHD1 to block HIV-1 but not SIV requires expression of MxB.Virology. 2019 May;531:260-268. doi: 10.1016/j.virol.2019.03.018. Epub 2019 Mar 30. Virology. 2019. PMID: 30959264 Free PMC article.
-
Karyopherin Alpha 2 Promotes the Inflammatory Response in Rat Pancreatic Acinar Cells Via Facilitating NF-κB Activation.Dig Dis Sci. 2016 Mar;61(3):747-57. doi: 10.1007/s10620-015-3948-6. Epub 2015 Nov 2. Dig Dis Sci. 2016. PMID: 26526450
-
Mechanistic Interplay between HIV-1 Reverse Transcriptase Enzyme Kinetics and Host SAMHD1 Protein: Viral Myeloid-Cell Tropism and Genomic Mutagenesis.Viruses. 2022 Jul 26;14(8):1622. doi: 10.3390/v14081622. Viruses. 2022. PMID: 35893688 Free PMC article. Review.
-
Insights from the Molecular Modelling and Docking Analysis of AIF-NLS complex to infer Nuclear Translocation of the Protein.Bioinformation. 2018 Mar 31;14(3):132-139. doi: 10.6026/97320630014132. eCollection 2018. Bioinformation. 2018. PMID: 29785072 Free PMC article.
References
-
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

