The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu
- PMID: 24711368
- PMCID: PMC4041430
- DOI: 10.1093/nar/gku256
The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu
Abstract
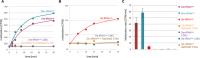
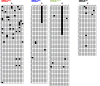
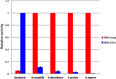
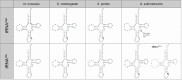
Dnmt2 enzymes are conserved in eukaryotes, where they methylate C38 of tRNA-Asp with high activity. Here, the activity of one of the very few prokaryotic Dnmt2 homologs from Geobacter species (GsDnmt2) was investigated. GsDnmt2 was observed to methylate tRNA-Asp from flies and mice. Unexpectedly, it had only a weak activity toward its matching Geobacter tRNA-Asp, but methylated Geobacter tRNA-Glu with good activity. In agreement with this result, we show that tRNA-Glu is methylated in Geobacter while the methylation is absent in tRNA-Asp. The activities of Dnmt2 enzymes from Homo sapiens, Drosophila melanogaster, Schizosaccharomyces pombe and Dictyostelium discoideum for methylation of the Geobacter tRNA-Asp and tRNA-Glu were determined showing that all these Dnmt2s preferentially methylate tRNA-Asp. Hence, the GsDnmt2 enzyme has a swapped transfer ribonucleic acid (tRNA) specificity. By comparing the different tRNAs, a characteristic sequence pattern was identified in the variable loop of all preferred tRNA substrates. An exchange of two nucleotides in the variable loop of murine tRNA-Asp converted it to the corresponding variable loop of tRNA-Glu and led to a strong reduction of GsDnmt2 activity. Interestingly, the same loss of activity was observed with human DNMT2, indicating that the variable loop functions as a specificity determinant in tRNA recognition of Dnmt2 enzymes.
© The Author(s) 2014. Published by Oxford University Press.
Figures



Similar articles
-
Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA).Nucleic Acids Res. 2013 Oct;41(18):8615-27. doi: 10.1093/nar/gkt634. Epub 2013 Jul 22. Nucleic Acids Res. 2013. PMID: 23877245 Free PMC article.
-
Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism.RNA. 2008 Aug;14(8):1663-70. doi: 10.1261/rna.970408. Epub 2008 Jun 20. RNA. 2008. PMID: 18567810 Free PMC article.
-
Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling.Nucleic Acids Res. 2012 Dec;40(22):11648-58. doi: 10.1093/nar/gks956. Epub 2012 Oct 15. Nucleic Acids Res. 2012. PMID: 23074192 Free PMC article.
-
Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification.Biomolecules. 2017 Feb 10;7(1):14. doi: 10.3390/biom7010014. Biomolecules. 2017. PMID: 28208632 Free PMC article. Review.
-
[Dnmt2 is the Most Evolutionary Conserved and Enigmatic Cytosine DNA Methyltransferase in Eukaryotes].Genetika. 2016 Mar;52(3):269-82. Genetika. 2016. PMID: 27281847 Review. Russian.
Cited by
-
The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA.RNA Biol. 2017 Sep 2;14(9):1241-1251. doi: 10.1080/15476286.2016.1236170. Epub 2016 Nov 7. RNA Biol. 2017. PMID: 27819523 Free PMC article.
-
Expression of KIAA1456 in lung cancer tissue and its effects on proliferation, migration and invasion of lung cancer cells.Oncol Lett. 2018 Sep;16(3):3791-3795. doi: 10.3892/ol.2018.9119. Epub 2018 Jul 10. Oncol Lett. 2018. PMID: 30127990 Free PMC article.
-
The identification and characterization of non-coding and coding RNAs and their modified nucleosides by mass spectrometry.RNA Biol. 2014;11(12):1568-85. doi: 10.4161/15476286.2014.992280. RNA Biol. 2014. PMID: 25616408 Free PMC article. Review.
-
Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine.Nucleic Acids Res. 2015 Dec 15;43(22):10952-62. doi: 10.1093/nar/gkv980. Epub 2015 Sep 30. Nucleic Acids Res. 2015. PMID: 26424849 Free PMC article.
-
To be or not to be modified: Miscellaneous aspects influencing nucleotide modifications in tRNAs.IUBMB Life. 2019 Aug;71(8):1126-1140. doi: 10.1002/iub.2041. Epub 2019 Apr 1. IUBMB Life. 2019. PMID: 30932315 Free PMC article. Review.
References
-
- Carell T., Brandmayr C., Hienzsch A., Muller M., Pearson D., Reiter V., Thoma I., Thumbs P., Wagner M. Structure and function of noncanonical nucleobases. Angew. Chem. Int. Ed. Engl. 2012;51:7110–7131. - PubMed
-
- El Yacoubi B., Bailly M., de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. - PubMed
-
- Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

