How human tumor viruses make use of autophagy
- PMID: 24710493
- PMCID: PMC3901112
- DOI: 10.3390/cells1030617
How human tumor viruses make use of autophagy
Abstract
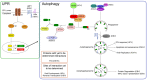
Viruses commandeer regulatory pathways of their hosts to optimize their success as cellular parasites. The human tumor viruses, Epstein-Barr Virus (EBV), Kaposi's Sarcoma Herpesvirus (KSHV), Hepatitis B Virus (HBV), and Hepatitis C Virus (HCV) all affect autophagy for their own ends. EBV and KSHV regulate it during latent infections, a phase when no progeny virus is produced, while HBV and HCV use autophagy to promote their productive infections. Here we shall compare and contrast how these human tumor viruses regulate autophagy and what they gain by the appropriation of this cellular pathway.
Figures
Similar articles
-
Regulation of Autophagy in Cells Infected With Oncogenic Human Viruses and Its Impact on Cancer Development.Front Cell Dev Biol. 2020 Feb 28;8:47. doi: 10.3389/fcell.2020.00047. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32181249 Free PMC article. Review.
-
Modulation of the autophagy pathway by human tumor viruses.Semin Cancer Biol. 2013 Oct;23(5):323-8. doi: 10.1016/j.semcancer.2013.05.005. Epub 2013 May 29. Semin Cancer Biol. 2013. PMID: 23727156 Free PMC article. Review.
-
Oncogenic Virus Infections in the General Population and End-stage Renal Disease Patients With Special Emphasis on Kaposi's Sarcoma Associated Herpes Virus (KSHV) in Northeast of Iran.Jundishapur J Microbiol. 2015 Mar 21;8(3):e14920. doi: 10.5812/jjm.14920. eCollection 2015 Mar. Jundishapur J Microbiol. 2015. PMID: 25834713 Free PMC article.
-
Mutual detection of Kaposi's sarcoma-associated herpesvirus and Epstein-Barr virus in blood and saliva of Cameroonians with and without Kaposi's sarcoma.Int J Cancer. 2019 Nov 1;145(9):2468-2477. doi: 10.1002/ijc.32546. Epub 2019 Jul 18. Int J Cancer. 2019. PMID: 31265124
-
Distinct patterns of viral antigen expression in Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus coinfected body-cavity-based lymphoma cell lines: potential switches in latent gene expression due to coinfection.Virology. 1999 Sep 15;262(1):18-30. doi: 10.1006/viro.1999.9876. Virology. 1999. PMID: 10489337
Cited by
-
ASPP2 inhibits hepatitis B virus replication by preventing nucleus translocation of HSF1 and attenuating the transactivation of ATG7.J Cell Mol Med. 2021 Jul;25(14):6899-6908. doi: 10.1111/jcmm.16699. Epub 2021 Jun 4. J Cell Mol Med. 2021. PMID: 34085409 Free PMC article.
-
Modulation of the unfolded protein response by the human hepatitis B virus.Front Microbiol. 2014 Aug 19;5:433. doi: 10.3389/fmicb.2014.00433. eCollection 2014. Front Microbiol. 2014. PMID: 25191311 Free PMC article. Review.
-
Novel Role of vBcl2 in the Virion Assembly of Kaposi's Sarcoma-Associated Herpesvirus.J Virol. 2018 Jan 30;92(4):e00914-17. doi: 10.1128/JVI.00914-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29167347 Free PMC article.
-
Epstein-Barr Functional Mimicry: Pathogenicity of Oncogenic Latent Membrane Protein-1 in Systemic Lupus Erythematosus and Autoimmunity.Front Immunol. 2021 Feb 3;11:606936. doi: 10.3389/fimmu.2020.606936. eCollection 2020. Front Immunol. 2021. PMID: 33613527 Free PMC article.
-
How Oncogenic Viruses Exploit p62-Mediated Selective Autophagy for Cancer Development.Ann Immunol Immunother. 2021;3(1):134. Epub 2021 Mar 5. Ann Immunol Immunother. 2021. PMID: 34632457 Free PMC article. No abstract available.
References
-
- Long H.M., Haigh T.A., Gudgeon N.H., Leen A.M., Tsang C.W., Brooks J., Landais E., Houssaint E., Lee S.P., Rickinson A.B., et al. CD4+ T-Cell responses to Epstein-Barr Virus (EBV) latent-cycle antigens and the recognition of EBV-transformed lymphoblastoid cell lines. J. Virol. 2005;79:4896–4907. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

