A smooth muscle-like origin for beige adipocytes
- PMID: 24709624
- PMCID: PMC4052772
- DOI: 10.1016/j.cmet.2014.03.025
A smooth muscle-like origin for beige adipocytes
Abstract
Thermogenic UCP1-positive cells, which include brown and beige adipocytes, transform chemical energy into heat and increase whole-body energy expenditure. Using a ribosomal profiling approach, we present a comprehensive molecular description of brown and beige gene expression from multiple fat depots in vivo. This UCP1-TRAP data set demonstrates striking similarities and important differences between these cell types, including a smooth muscle-like signature expressed by beige, but not classical brown, adipocytes. In vivo fate mapping using either a constitutive or an inducible Myh11-driven Cre demonstrates that at least a subset of beige cells arise from a smooth muscle-like origin. Finally, ectopic expression of PRDM16 converts bona fide vascular smooth muscle cells into Ucp1-positive adipocytes in vitro. These results establish a portrait of brown and beige adipocyte gene expression in vivo and identify a smooth muscle-like origin for beige cells.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
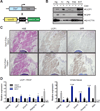



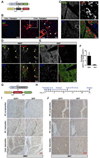
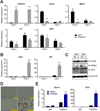
Comment in
-
Fat or fiction: origins matter.Cell Metab. 2014 Jun 3;19(6):900-1. doi: 10.1016/j.cmet.2014.05.007. Cell Metab. 2014. PMID: 24896537
Similar articles
-
Functional thermogenic beige adipogenesis is inducible in human neck fat.Int J Obes (Lond). 2014 Feb;38(2):170-6. doi: 10.1038/ijo.2013.82. Epub 2013 May 21. Int J Obes (Lond). 2014. PMID: 23736373 Free PMC article.
-
Brite/beige fat and UCP1 - is it thermogenesis?Biochim Biophys Acta. 2014 Jul;1837(7):1075-82. doi: 10.1016/j.bbabio.2014.02.008. Epub 2014 Feb 14. Biochim Biophys Acta. 2014. PMID: 24530356
-
Thermogenic ability of uncoupling protein 1 in beige adipocytes in mice.PLoS One. 2013 Dec 30;8(12):e84229. doi: 10.1371/journal.pone.0084229. eCollection 2013. PLoS One. 2013. PMID: 24386355 Free PMC article.
-
Adaptive thermogenesis in adipocytes: is beige the new brown?Genes Dev. 2013 Feb 1;27(3):234-50. doi: 10.1101/gad.211649.112. Genes Dev. 2013. PMID: 23388824 Free PMC article. Review.
-
Role of PRDM16 in the activation of brown fat programming. Relevance to the development of obesity.Histol Histopathol. 2013 Nov;28(11):1411-25. doi: 10.14670/HH-28.1411. Epub 2013 Jun 17. Histol Histopathol. 2013. PMID: 23771475 Review.
Cited by
-
GQ-16, a TZD-Derived Partial PPARγ Agonist, Induces the Expression of Thermogenesis-Related Genes in Brown Fat and Visceral White Fat and Decreases Visceral Adiposity in Obese and Hyperglycemic Mice.PLoS One. 2016 May 3;11(5):e0154310. doi: 10.1371/journal.pone.0154310. eCollection 2016. PLoS One. 2016. PMID: 27138164 Free PMC article.
-
Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation.Genes Dev. 2016 Aug 15;30(16):1822-36. doi: 10.1101/gad.285312.116. Epub 2016 Aug 26. Genes Dev. 2016. PMID: 27566776 Free PMC article.
-
Metabolic Improvement via Enhancing Thermogenic Fat-Mediated Non-shivering Thermogenesis: From Rodents to Humans.Front Endocrinol (Lausanne). 2020 Sep 10;11:633. doi: 10.3389/fendo.2020.00633. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33013706 Free PMC article. Review.
-
Beige Adipose Tissue Identification and Marker Specificity-Overview.Front Endocrinol (Lausanne). 2021 Mar 12;12:599134. doi: 10.3389/fendo.2021.599134. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 33776911 Free PMC article. Review.
-
Sympathetic inputs regulate adaptive thermogenesis in brown adipose tissue through cAMP-Salt inducible kinase axis.Sci Rep. 2018 Jul 20;8(1):11001. doi: 10.1038/s41598-018-29333-6. Sci Rep. 2018. PMID: 30030465 Free PMC article.
References
-
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological reviews. 2004;84:277–359. - PubMed
-
- Cederberg A, Gronning LM, Ahren B, Tasken K, Carlsson P, Enerback S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

