Sumoylation differentially regulates Sp1 to control cell differentiation
- PMID: 24706897
- PMCID: PMC3992630
- DOI: 10.1073/pnas.1315034111
Sumoylation differentially regulates Sp1 to control cell differentiation
Abstract
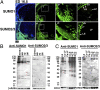
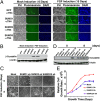
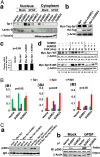
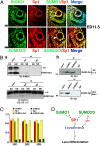
The mammalian small ubiquitin-like modifiers (SUMOs) are actively involved in regulating differentiation of different cell types. However, the functional differences between SUMO isoforms and their mechanisms of action remain largely unknown. Using the ocular lens as a model system, we demonstrate that different SUMOs display distinct functions in regulating differentiation of epithelial cells into fiber cells. During lens differentiation, SUMO1 and SUMO2/3 displayed different expression, localization, and targets, suggesting differential functions. Indeed, overexpression of SUMO2/3, but not SUMO1, inhibited basic (b) FGF-induced cell differentiation. In contrast, knockdown of SUMO1, but not SUMO2/3, also inhibited bFGF action. Mechanistically, specificity protein 1 (Sp1), a major transcription factor that controls expression of lens-specific genes such as β-crystallins, was positively regulated by SUMO1 but negatively regulated by SUMO2. SUMO2 was found to inhibit Sp1 functions through several mechanisms: sumoylating it at K683 to attenuate DNA binding, and at K16 to increase its turnover. SUMO2 also interfered with the interaction between Sp1 and the coactivator, p300, and recruited a repressor, Sp3 to β-crystallin gene promoters, to negatively regulate their expression. Thus, stable SUMO1, but diminishing SUMO2/3, during lens development is necessary for normal lens differentiation. In support of this conclusion, SUMO1 and Sp1 formed complexes during early and later stages of lens development. In contrast, an interaction between SUMO2/3 and Sp1 was detected only during the initial lens vesicle stage. Together, our results establish distinct roles of different SUMO isoforms and demonstrate for the first time, to our knowledge, that Sp1 acts as a major transcription factor target for SUMO control of cell differentiation.
Keywords: crystallin gene expression; eye development; transcription regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
SENP3 regulates the global protein turnover and the Sp1 level via antagonizing SUMO2/3-targeted ubiquitination and degradation.Protein Cell. 2016 Jan;7(1):63-77. doi: 10.1007/s13238-015-0216-7. Epub 2015 Oct 28. Protein Cell. 2016. PMID: 26511642 Free PMC article.
-
Spatiotemporal distribution of SUMOylation components during mouse brain development.J Comp Neurol. 2014 Sep 1;522(13):3020-36. doi: 10.1002/cne.23563. J Comp Neurol. 2014. PMID: 24639124
-
HIF-1α SUMOylation affects the stability and transcriptional activity of HIF-1α in human lens epithelial cells.Graefes Arch Clin Exp Ophthalmol. 2015 Aug;253(8):1279-90. doi: 10.1007/s00417-015-2999-x. Epub 2015 Apr 16. Graefes Arch Clin Exp Ophthalmol. 2015. PMID: 25877955
-
Paralogue-Specific Roles of SUMO1 and SUMO2/3 in Protein Quality Control and Associated Diseases.Cells. 2023 Dec 20;13(1):8. doi: 10.3390/cells13010008. Cells. 2023. PMID: 38201212 Free PMC article. Review.
-
The post-translational modification, SUMOylation, and cancer (Review).Int J Oncol. 2018 Apr;52(4):1081-1094. doi: 10.3892/ijo.2018.4280. Epub 2018 Feb 22. Int J Oncol. 2018. PMID: 29484374 Free PMC article. Review.
Cited by
-
The requirement of SUMO2/3 for SENP2 mediated extraembryonic and embryonic development.Dev Dyn. 2020 Feb;249(2):237-244. doi: 10.1002/dvdy.125. Epub 2019 Nov 1. Dev Dyn. 2020. PMID: 31625212 Free PMC article.
-
Plasma amino acids profile in first-episode psychosis, unaffected siblings and community-based controls.Sci Rep. 2020 Dec 8;10(1):21423. doi: 10.1038/s41598-020-78559-w. Sci Rep. 2020. PMID: 33293633 Free PMC article.
-
Regulation of transcription factors by sumoylation.Transcription. 2017 Aug 8;8(4):220-231. doi: 10.1080/21541264.2017.1311829. Epub 2017 Apr 5. Transcription. 2017. PMID: 28379052 Free PMC article. Review.
-
SUMO2 regulates vascular endothelial function and oxidative stress in mice.Am J Physiol Heart Circ Physiol. 2019 Dec 1;317(6):H1292-H1300. doi: 10.1152/ajpheart.00530.2019. Epub 2019 Oct 4. Am J Physiol Heart Circ Physiol. 2019. PMID: 31584834 Free PMC article.
-
Editorial: Apoptosis and Senescence in Vertebrate Development.Front Cell Dev Biol. 2022 Jan 5;9:834517. doi: 10.3389/fcell.2021.834517. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35071249 Free PMC article. No abstract available.
References
-
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol. 2007;8(12):947–956. - PubMed
-
- Lapenta V, et al. SMT3A, a human homologue of the S. cerevisiae SMT3 gene, maps to chromosome 21qter and defines a novel gene family. Genomics. 1997;40(2):362–366. - PubMed
-
- Kamitani T, Kito K, Nguyen HP, Fukuda-Kamitani T, Yeh ET. Characterization of a second member of the sentrin family of ubiquitin-like proteins. J Biol Chem. 1998;273(18):11349–11353. - PubMed
-
- Vertegaal AC, et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5(12):2298–2310. - PubMed
-
- Alkuraya FS, et al. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313(5794):1751. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

