Template-directed chemical ligation to obtain 3'-3' and 5'-5' phosphodiester DNA linkages
- PMID: 24699719
- PMCID: PMC3975322
- DOI: 10.1038/srep04595
Template-directed chemical ligation to obtain 3'-3' and 5'-5' phosphodiester DNA linkages
Abstract
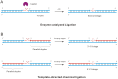
Up to now, the direct ligation of two DNA fragments with opposite directions to obtain 3'-3' or 5'-5' phosphate ester bonds is still challenging. The only way to obtain DNA oligonucleotides containing a 3'-3' or 5'-5' inversion of polarity sites is based on professional DNA chemical synthesis. Herein, we demonstrate a convenient template-directed chemical ligation that enables 3'-3' and 5'-5' linkages of two DNA oligonucleotides. This method is based on the assembly of two oligonucleotides on a template in opposite directions through forming antiparallel and parallel duplexes simultaneously, followed by coupling with N-Cyanoimidazole under mild condition. Moreover, on the basis of DNA oligonucleotides with 5'-5' linkage obtained through our template-directed chemical ligation, we developed a new cDNA display technique for in vitro selection of functional polypeptides.
Figures



Similar articles
-
Template-dependent DNA ligation for the synthesis of modified oligonucleotides.Nat Commun. 2024 Sep 13;15(1):8009. doi: 10.1038/s41467-024-52141-8. Nat Commun. 2024. PMID: 39271668 Free PMC article.
-
Nonenzymatic, template-directed ligation of oligoribonucleotides is highly regioselective for the formation of 3'-5' phosphodiester bonds.J Am Chem Soc. 1996 Apr 10;118(14):3340-4. doi: 10.1021/ja9537134. J Am Chem Soc. 1996. PMID: 11539268
-
Canonical nucleosides can be utilized by T4 DNA ligase as universal template bases at ligation junctions.Nucleic Acids Res. 2003 Jun 15;31(12):3208-16. doi: 10.1093/nar/gkg415. Nucleic Acids Res. 2003. PMID: 12799448 Free PMC article.
-
Click nucleic acid ligation: applications in biology and nanotechnology.Acc Chem Res. 2012 Aug 21;45(8):1258-67. doi: 10.1021/ar200321n. Epub 2012 Mar 22. Acc Chem Res. 2012. PMID: 22439702 Free PMC article. Review.
-
Chemical ligation methods for the tagging of DNA-encoded chemical libraries.Curr Opin Chem Biol. 2015 Jun;26:80-8. doi: 10.1016/j.cbpa.2015.02.015. Epub 2015 Mar 8. Curr Opin Chem Biol. 2015. PMID: 25756406 Review.
Cited by
-
Rapid Chemical Ligation of DNA and Acyclic Threoninol Nucleic Acid (aTNA) for Effective Nonenzymatic Primer Extension.J Am Chem Soc. 2023 Aug 16;145(32):17872-17880. doi: 10.1021/jacs.3c04979. Epub 2023 Jul 19. J Am Chem Soc. 2023. PMID: 37466125 Free PMC article.
-
Common and Potentially Prebiotic Origin for Precursors of Nucleotide Synthesis and Activation.J Am Chem Soc. 2017 Jul 5;139(26):8780-8783. doi: 10.1021/jacs.7b01562. Epub 2017 Jun 22. J Am Chem Soc. 2017. PMID: 28640999 Free PMC article.
-
Nonenzymatic polymerase-like template-directed synthesis of acyclic L-threoninol nucleic acid.Nat Commun. 2021 Feb 5;12(1):804. doi: 10.1038/s41467-021-21128-0. Nat Commun. 2021. PMID: 33547322 Free PMC article.
References
-
- Li T. & Nicolaou K. C. Chemical self-replication of palindromic duplex DNA. Nature 369, 218–221 (1994). - PubMed
-
- Vonkiedrowski G. A self-replicating hexadeoxynucleotide. Angew. Chem. Int. Ed. Engl. 25, 932–935 (1986).
-
- Xu L. J. & Liu D. S. Functional evolution on the assembled DNA template. Chem. Soc. Rev. 39, 150–155 (2010). - PubMed
-
- Vandesande J. H. et al. Parallel stranded DNA. Science 241, 551–557 (1988). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

