The carboxy terminal region of the human cytomegalovirus immediate early 1 (IE1) protein disrupts type II inteferon signaling
- PMID: 24699362
- PMCID: PMC4014707
- DOI: 10.3390/v6041502
The carboxy terminal region of the human cytomegalovirus immediate early 1 (IE1) protein disrupts type II inteferon signaling
Abstract
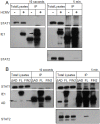
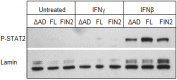
Interferons (IFNs) activate the first lines of defense against viruses, and promote innate and adaptive immune responses to viruses. We report that the immediate early 1 (IE1) protein of human cytomegalovirus (HCMV) disrupts signaling by IFNγ. The carboxyl-terminal region of IE1 is required for this function. We found no defect in the initial events in IFNγ signaling or in nuclear accumulation of signal transducer and activator of transcription 1 (STAT1) in IE1-expressing cells. Moreover, we did not observe an association between disruption of IFNγ signaling and nuclear domain 10 (ND10) disruption. However, there is reduced binding of STAT1 homodimers to target gamma activated sequence (GAS) elements in the presence of IE1. Co-immunoprecipitation studies failed to support a direct interaction between IE1 and STAT1, although these studies revealed that the C-terminal region of IE1 was required for interaction with STAT2. Together, these results indicate that IE1 disrupts IFNγ signaling by interfering with signaling events in the nucleus through a novel mechanism.
Figures


Similar articles
-
Human Cytomegalovirus Immediate-Early 1 Protein Rewires Upstream STAT3 to Downstream STAT1 Signaling Switching an IL6-Type to an IFNγ-Like Response.PLoS Pathog. 2016 Jul 7;12(7):e1005748. doi: 10.1371/journal.ppat.1005748. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27387064 Free PMC article.
-
Characterization of Recombinant Human Cytomegaloviruses Encoding IE1 Mutants L174P and 1-382 Reveals that Viral Targeting of PML Bodies Perturbs both Intrinsic and Innate Immune Responses.J Virol. 2015 Nov 11;90(3):1190-205. doi: 10.1128/JVI.01973-15. Print 2016 Feb 1. J Virol. 2015. PMID: 26559840 Free PMC article.
-
Human cytomegalovirus IE1 protein elicits a type II interferon-like host cell response that depends on activated STAT1 but not interferon-γ.PLoS Pathog. 2011 Apr;7(4):e1002016. doi: 10.1371/journal.ppat.1002016. Epub 2011 Apr 14. PLoS Pathog. 2011. PMID: 21533215 Free PMC article.
-
The Human CMV IE1 Protein: An Offender of PML Nuclear Bodies.Adv Anat Embryol Cell Biol. 2017;223:77-94. doi: 10.1007/978-3-319-53168-7_4. Adv Anat Embryol Cell Biol. 2017. PMID: 28528440 Review.
-
Intrinsic Immune Mechanisms Restricting Human Cytomegalovirus Replication.Viruses. 2021 Jan 26;13(2):179. doi: 10.3390/v13020179. Viruses. 2021. PMID: 33530304 Free PMC article. Review.
Cited by
-
Bright and Early: Inhibiting Human Cytomegalovirus by Targeting Major Immediate-Early Gene Expression or Protein Function.Viruses. 2020 Jan 16;12(1):110. doi: 10.3390/v12010110. Viruses. 2020. PMID: 31963209 Free PMC article. Review.
-
The essential role of guinea pig cytomegalovirus (GPCMV) IE1 and IE2 homologs in viral replication and IE1-mediated ND10 targeting.Virology. 2017 Apr;504:122-140. doi: 10.1016/j.virol.2017.01.023. Epub 2017 Feb 10. Virology. 2017. PMID: 28189970 Free PMC article.
-
Human Cytomegalovirus Immediate-Early 1 Protein Rewires Upstream STAT3 to Downstream STAT1 Signaling Switching an IL6-Type to an IFNγ-Like Response.PLoS Pathog. 2016 Jul 7;12(7):e1005748. doi: 10.1371/journal.ppat.1005748. eCollection 2016 Jul. PLoS Pathog. 2016. PMID: 27387064 Free PMC article.
-
Who's Driving? Human Cytomegalovirus, Interferon, and NFκB Signaling.Viruses. 2018 Aug 21;10(9):447. doi: 10.3390/v10090447. Viruses. 2018. PMID: 30134546 Free PMC article. Review.
-
Foot-and-mouth disease virus structural protein VP3 degrades Janus kinase 1 to inhibit IFN-γ signal transduction pathways.Cell Cycle. 2016;15(6):850-60. doi: 10.1080/15384101.2016.1151584. Cell Cycle. 2016. PMID: 26901336 Free PMC article.
References
-
- Britt W.J., Alford C. A Cytomegalovirus. In: Fields B., Knipe D.M., Howley P.M., editors. Fields Virology. Volume 2. Lippincott-Raven; Philadelphia, PA, USA: 1996. pp. 2493–2523.
-
- Goodbourn S., Didcock L., Randall R.E. Interferons: Cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 2000;81:2341–2364. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

