A novel, non-radioactive eukaryotic in vitro transcription assay for sensitive quantification of RNA polymerase II activity
- PMID: 24694320
- PMCID: PMC4021065
- DOI: 10.1186/1471-2199-15-7
A novel, non-radioactive eukaryotic in vitro transcription assay for sensitive quantification of RNA polymerase II activity
Abstract
Background: Many studies of the eukaryotic transcription mechanism and its regulation rely on in vitro assays. Conventional RNA polymerase II transcription assays are based on radioactive labelling of the newly synthesized RNA. Due to the inefficient in vitro transcription, the detection of the RNA involving purification and gel electrophoresis is laborious and not always quantitative.
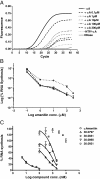
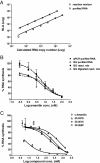
Results: Herein, we describe a new, non-radioactive, robust and reproducible eukaryotic in vitro transcription assay that has been established in our laboratory. Upon transcription, the newly synthesized RNA is directly detected and quantified using the QuantiGene assay. Alternatively, the RNA can be purified and a primer extension followed by PCR detection or qPCR quantification can be performed. When applied to assess the activity of RNA polymerase II inhibitors, this new method allowed an accurate estimation of their relative potency.
Conclusions: Our novel assay provides a non-radioactive alternative to a standard in vitro transcription assay that allows for sensitive detection and precise quantification of the newly transcribed, unlabelled RNA and is particularly useful for quantification of strong transcriptional inhibitors like α-amanitin. Moreover, the method can be easily adapted to quantify the reaction yield and the transcription efficiency of other eukaryotic in vitro systems, thus providing a complementary tool for the field of transcriptional research.
Figures



Similar articles
-
Establishment and Validation of a Non-Radioactive Method for In Vitro Transcription Assay Using Primer Extension and Quantitative Real Time PCR.PLoS One. 2015 Aug 7;10(8):e0135317. doi: 10.1371/journal.pone.0135317. eCollection 2015. PLoS One. 2015. PMID: 26252791 Free PMC article.
-
Transcription in the early diverging eukaryote Trichomonas vaginalis: an unusual RNA polymerase II and alpha-amanitin-resistant transcription of protein-coding genes.J Mol Evol. 1996 Sep;43(3):253-62. doi: 10.1007/BF02338833. J Mol Evol. 1996. PMID: 8703091
-
Assays for investigating transcription by RNA polymerase II in vitro.Methods. 1997 Jul;12(3):192-202. doi: 10.1006/meth.1997.0471. Methods. 1997. PMID: 9237163 Review.
-
Selective in vitro transcription by purified yeast RNA polymerase II on cloned 2 micron DNA.Nucleic Acids Res. 1981 Aug 25;9(16):3959-78. doi: 10.1093/nar/9.16.3959. Nucleic Acids Res. 1981. PMID: 7029462 Free PMC article.
-
Overlapping pathways dictate termination of RNA polymerase II transcription.Biochimie. 2007 Oct;89(10):1177-82. doi: 10.1016/j.biochi.2007.05.007. Epub 2007 Jun 2. Biochimie. 2007. PMID: 17629387 Review.
Cited by
-
Revisiting T7 RNA polymerase transcription in vitro with the Broccoli RNA aptamer as a simplified real-time fluorescent reporter.J Biol Chem. 2021 Jan-Jun;296:100175. doi: 10.1074/jbc.RA120.014553. Epub 2020 Dec 16. J Biol Chem. 2021. PMID: 33303627 Free PMC article.
-
Establishment and Validation of a Non-Radioactive Method for In Vitro Transcription Assay Using Primer Extension and Quantitative Real Time PCR.PLoS One. 2015 Aug 7;10(8):e0135317. doi: 10.1371/journal.pone.0135317. eCollection 2015. PLoS One. 2015. PMID: 26252791 Free PMC article.
-
Isolation of Active Caenorhabditis elegans Nuclear Extract and Reconstitution for In Vitro Transcription.J Vis Exp. 2021 Aug 11;(174):10.3791/62723. doi: 10.3791/62723. J Vis Exp. 2021. PMID: 34459806 Free PMC article.
-
Heterozygous deletion of chromosome 17p renders prostate cancer vulnerable to inhibition of RNA polymerase II.Nat Commun. 2018 Oct 22;9(1):4394. doi: 10.1038/s41467-018-06811-z. Nat Commun. 2018. PMID: 30349055 Free PMC article.
-
A novel in vitro Caenorhabditis elegans transcription system.BMC Mol Cell Biol. 2020 Nov 30;21(1):87. doi: 10.1186/s12860-020-00332-8. BMC Mol Cell Biol. 2020. PMID: 33256604 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

