In silico investigation of pH-dependence of prolactin and human growth hormone binding to human prolactin receptor
- PMID: 24683423
- PMCID: PMC3966486
- DOI: 10.4208/cicp.170911.131011s
In silico investigation of pH-dependence of prolactin and human growth hormone binding to human prolactin receptor
Abstract
Experimental data shows that the binding of human prolactin (hPRL) to human prolactin receptor (hPRLr-ECD) is strongly pH-dependent, while the binding of the same receptor to human growth hormone (hGH) is pH-independent. Here we carry in silico analysis of the molecular effects causing such a difference and reveal the role of individual amino acids. It is shown that the computational modeling correctly predicts experimentally determined pKa's of histidine residues in an unbound state in the majority of the cases and the pH-dependence of the binding free energy. Structural analysis carried in conjunction with calculated pH-dependence of the binding revealed that the main reason for pH-dependence of the binding of hPRL-hPRLr-ECD is a number of salt- bridges across the interface of the complex, while no salt-bridges are formed in the hGH-hPRlr-ECD. Specifically, most of the salt-bridges involve histidine residues and this is the reason for the pH-dependence across a physiological range of pH. The analysis not only revealed the molecular mechanism of the pH-dependence of the hPRL-hPRLr-ECD, but also provided critical insight into the underlying physic-chemical mechanism.
Keywords: electrostatics; human growth hormone; human prolactin; human prolactin receptor; pH-dependence; pKa calculations.
Figures


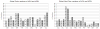
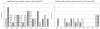

Similar articles
-
The kinetics of binding human prolactin, but not growth hormone, to the prolactin receptor vary over a physiologic pH range.Biochemistry. 2007 Mar 6;46(9):2398-410. doi: 10.1021/bi061958v. Epub 2007 Feb 6. Biochemistry. 2007. PMID: 17279774
-
Novel reagents for human prolactin research: large-scale preparation and characterization of prolactin receptor extracellular domain, non-pegylated and pegylated prolactin and prolactin receptor antagonist.Protein Eng Des Sel. 2018 Jan 1;31(1):7-16. doi: 10.1093/protein/gzx062. Protein Eng Des Sel. 2018. PMID: 29281090
-
Contribution of individual histidines to the global stability of human prolactin.Protein Sci. 2009 May;18(5):909-20. doi: 10.1002/pro.100. Protein Sci. 2009. PMID: 19384991 Free PMC article.
-
Comparison of the intermediate complexes of human growth hormone bound to the human growth hormone and prolactin receptors.Protein Sci. 1994 Oct;3(10):1697-705. doi: 10.1002/pro.5560031008. Protein Sci. 1994. PMID: 7849586 Free PMC article. Review.
-
The role of protonation states in ligand-receptor recognition and binding.Curr Pharm Des. 2013;19(23):4182-90. doi: 10.2174/1381612811319230004. Curr Pharm Des. 2013. PMID: 23170880 Free PMC article. Review.
Cited by
-
On the energy components governing molecular recognition in the framework of continuum approaches.Front Mol Biosci. 2015 Mar 6;2:5. doi: 10.3389/fmolb.2015.00005. eCollection 2015. Front Mol Biosci. 2015. PMID: 25988173 Free PMC article. Review.
-
Using a comprehensive approach to investigate the interaction between Kinesin-5/Eg5 and the microtubule.Comput Struct Biotechnol J. 2022 Aug 11;20:4305-4314. doi: 10.1016/j.csbj.2022.08.020. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36051882 Free PMC article.
-
Protonation and pK changes in protein-ligand binding.Q Rev Biophys. 2013 May;46(2):181-209. doi: 10.1017/S0033583513000024. Q Rev Biophys. 2013. PMID: 23889892 Free PMC article. Review.
-
Computation of pH-dependent binding free energies.Biopolymers. 2016 Jan;105(1):43-9. doi: 10.1002/bip.22702. Biopolymers. 2016. PMID: 26202905 Free PMC article. Review.
-
Electrostatic features for nucleocapsid proteins of SARS-CoV and SARS-CoV-2.Math Biosci Eng. 2021 Mar 9;18(3):2372-2383. doi: 10.3934/mbe.2021120. Math Biosci Eng. 2021. PMID: 33892550 Free PMC article.
References
-
- Alexov E. Protein-protein interactions. Curr Pharm Biotechnol. 2008;9(2):55–6. - PubMed
-
- Alexov E. Calculating proton uptake/release and binding free energy taking into account ionization and conformation changes induced by protein-inhibitor association: application to plasmepsin, cathepsin D and endothiapepsin-pepstatin complexes. Proteins. 2004;56(3):572–84. - PubMed
-
- Bevilacqua PC, Brown TS, Chadalavada D, Lecomte J, Moody E, Nakano SI. Linkage between proton binding and folding in RNA:implications for RNA catalysis. Biochem Soc Trans. 2005;33(Pt 3):466–70. - PubMed
-
- Matthew JB, Gurd FR, Garcia-Moreno B, Flanagan MA, March KL, Shire SJ. pH-dependent processes in proteins. CRC Crit Rev Biochem. 1985;18(2):91–197. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
