A tRNA body with high affinity for EF-Tu hastens ribosomal incorporation of unnatural amino acids
- PMID: 24671767
- PMCID: PMC3988565
- DOI: 10.1261/rna.042234.113
A tRNA body with high affinity for EF-Tu hastens ribosomal incorporation of unnatural amino acids
Abstract
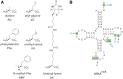
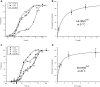
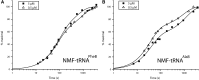
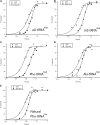
There is evidence that tRNA bodies have evolved to reduce differences between aminoacyl-tRNAs in their affinity to EF-Tu. Here, we study the kinetics of incorporation of L-amino acids (AAs) Phe, Ala allyl-glycine (aG), methyl-serine (mS), and biotinyl-lysine (bK) using a tRNA(Ala)-based body (tRNA(AlaB)) with a high affinity for EF-Tu. Results are compared with previous data on the kinetics of incorporation of the same AAs using a tRNA(PheB) body with a comparatively low affinity for EF-Tu. All incorporations exhibited fast and slow phases, reflecting the equilibrium fraction of AA-tRNA in active ternary complex with EF-Tu:GTP before the incorporation reaction. Increasing the concentration of EF-Tu increased the amplitude of the fast phase and left its rate unaltered. This allowed estimation of the affinity of each AA-tRNA to EF-Tu:GTP during translation, showing about a 10-fold higher EF-Tu affinity for AA-tRNAs formed from the tRNA(AlaB) body than from the tRNA(PheB) body. At ∼1 µM EF-Tu, tRNA(AlaB) conferred considerably faster incorporation kinetics than tRNA(PheB), especially in the case of the bulky bK. In contrast, the swap to the tRNA(AlaB) body did not increase the fast phase fraction of N-methyl-Phe incorporation, suggesting that the slow incorporation of N-methyl-Phe had a different cause than low EF-Tu:GTP affinity. The total time for AA-tRNA release from EF-Tu:GDP, accommodation, and peptidyl transfer on the ribosome was similar for the tRNA(AlaB) and tRNA(PheB) bodies. We conclude that a tRNA body with high EF-Tu affinity can greatly improve incorporation of unnatural AAs in a potentially generalizable manner.
Keywords: EF-Tu affinity; ribosome; tRNAAla; translation kinetics; unnatural amino acid.
Figures



Similar articles
-
Inefficient delivery but fast peptide bond formation of unnatural L-aminoacyl-tRNAs in translation.J Am Chem Soc. 2012 Oct 31;134(43):17955-62. doi: 10.1021/ja3063524. Epub 2012 Oct 22. J Am Chem Soc. 2012. PMID: 23057558
-
Elongation factor-Tu can repetitively engage aminoacyl-tRNA within the ribosome during the proofreading stage of tRNA selection.Proc Natl Acad Sci U S A. 2020 Feb 18;117(7):3610-3620. doi: 10.1073/pnas.1904469117. Epub 2020 Feb 5. Proc Natl Acad Sci U S A. 2020. PMID: 32024753 Free PMC article.
-
Fluorescence characterization of the interaction of various transfer RNA species with elongation factor Tu.GTP: evidence for a new functional role for elongation factor Tu in protein biosynthesis.Biochemistry. 1990 May 8;29(18):4268-77. doi: 10.1021/bi00470a002. Biochemistry. 1990. PMID: 2190631
-
Elongation factor Tu, a GTPase triggered by codon recognition on the ribosome: mechanism and GTP consumption.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):1221-7. doi: 10.1139/o95-132. Biochem Cell Biol. 1995. PMID: 8722040 Review.
-
In Vitro Genetic Code Reprogramming for the Expansion of Usable Noncanonical Amino Acids.Annu Rev Biochem. 2022 Jun 21;91:221-243. doi: 10.1146/annurev-biochem-040320-103817. Annu Rev Biochem. 2022. PMID: 35729073 Review.
Cited by
-
Efficient Reassignment of a Frequent Serine Codon in Wild-Type Escherichia coli.ACS Synth Biol. 2016 Feb 19;5(2):163-71. doi: 10.1021/acssynbio.5b00197. Epub 2015 Nov 20. ACS Synth Biol. 2016. PMID: 26544153 Free PMC article.
-
β-Amino Acids Reduce Ternary Complex Stability and Alter the Translation Elongation Mechanism.ACS Cent Sci. 2024 Jun 4;10(6):1262-1275. doi: 10.1021/acscentsci.4c00314. eCollection 2024 Jun 26. ACS Cent Sci. 2024. PMID: 38947208 Free PMC article.
-
Extensive breaking of genetic code degeneracy with non-canonical amino acids.Nat Commun. 2023 Aug 17;14(1):5008. doi: 10.1038/s41467-023-40529-x. Nat Commun. 2023. PMID: 37591858 Free PMC article.
-
The central role of tRNA in genetic code expansion.Biochim Biophys Acta Gen Subj. 2017 Nov;1861(11 Pt B):3001-3008. doi: 10.1016/j.bbagen.2017.03.012. Epub 2017 Mar 18. Biochim Biophys Acta Gen Subj. 2017. PMID: 28323071 Free PMC article. Review.
-
Tuning tRNAs for improved translation.Front Genet. 2024 Jun 25;15:1436860. doi: 10.3389/fgene.2024.1436860. eCollection 2024. Front Genet. 2024. PMID: 38983271 Free PMC article. Review.
References
-
- Bain JD, Glabe CG, Dix TA, Chamberlin AR, Diala ES 1989. Biosynthetic site-specific incorporation of a non-natural amino acid into a polypeptide. J Am Chem Soc 111: 8013–8014
-
- Bieling P, Beringer M, Adio S, Rodnina MV 2006. Peptide bond formation does not involve acid-base catalysis by ribosomal residues. Nat Struct Mol Biol 13: 423–428 - PubMed
-
- Cload ST, Liu DR, Froland WA, Schultz PG 1996. Development of improved tRNAs for in vitro biosynthesis of proteins containing unnatural amino acids. Chem Biol 3: 1033–1038 - PubMed
-
- Doi Y, Ohtsuki T, Shimizu Y, Ueda T, Sisido M 2007. Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. J Am Chem Soc 129: 14458–14462 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
