Rad4 mainly functions in Chk1-mediated DNA damage checkpoint pathway as a scaffold protein in the fission yeast Schizosaccharomyces pombe
- PMID: 24663817
- PMCID: PMC3963969
- DOI: 10.1371/journal.pone.0092936
Rad4 mainly functions in Chk1-mediated DNA damage checkpoint pathway as a scaffold protein in the fission yeast Schizosaccharomyces pombe
Erratum in
-
Correction: Rad4 Mainly Functions in Chk1-Mediated DNA Damage Checkpoint Pathway as a Scaffold Protein in the Fission Yeast Schizosaccharomyces pombe.PLoS One. 2018 Sep 11;13(9):e0204070. doi: 10.1371/journal.pone.0204070. eCollection 2018. PLoS One. 2018. PMID: 30204808 Free PMC article.
Abstract
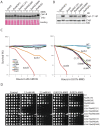
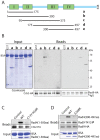
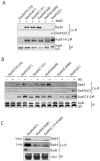
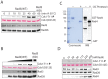
Rad4/Cut5 is a scaffold protein in the Chk1-mediated DNA damage checkpoint in S. pombe. However, whether it contains a robust ATR-activation domain (AAD) required for checkpoint signaling like its orthologs TopBP1 in humans and Dpb11 in budding yeast has been incompletely clear. To identify the putative AAD in Rad4, we carried out an extensive genetic screen looking for novel mutants with an enhanced sensitivity to replication stress or DNA damage in which the function of the AAD can be eliminated by the mutations. Two new mutations near the N-terminus were identified that caused significantly higher sensitivities to DNA damage or chronic replication stress than all previously reported mutants, suggesting that most of the checkpoint function of the protein is eliminated. However, these mutations did not affect the activation of Rad3 (ATR in humans) yet eliminated the scaffolding function of the protein required for the activation of Chk1. Several mutations were also identified in or near the recently reported AAD in the C-terminus of Rad4. However, all mutations in the C-terminus only slightly sensitized the cells to DNA damage. Interestingly, a mutant lacking the whole C-terminus was found resistant to DNA damage and replication stress almost like the wild type cells. Consistent with the resistance, all known Rad3 dependent phosphorylations of checkpoint proteins remained intact in the C-terminal deletion mutant, indicating that unlike that in Dpb11, the C-terminus of Rad4 does not contain a robust AAD. These results, together with those from the biochemical studies, show that Rad4 mainly functions as a scaffold protein in the Chk1, not the Cds1(CHK2 in humans), checkpoint pathway. It plays a minor role or is functionally redundant with an unknown factor in Rad3 activation.
Conflict of interest statement
Figures


Similar articles
-
The Rad4(TopBP1) ATR-activation domain functions in G1/S phase in a chromatin-dependent manner.PLoS Genet. 2012 Jun;8(6):e1002801. doi: 10.1371/journal.pgen.1002801. Epub 2012 Jun 28. PLoS Genet. 2012. PMID: 22761595 Free PMC article.
-
Delineating the position of rad4+/cut5+ within the DNA-structure checkpoint pathways in Schizosaccharomyces pombe.J Cell Sci. 2003 Sep 1;116(Pt 17):3519-29. doi: 10.1242/jcs.00677. Epub 2003 Jul 15. J Cell Sci. 2003. PMID: 12865439
-
Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1.Genes Dev. 2004 May 15;18(10):1154-64. doi: 10.1101/gad.291104. Genes Dev. 2004. PMID: 15155581 Free PMC article.
-
Rad3 and Sty1 function in Schizosaccharomyces pombe: an integrated response to DNA damage and environmental stress?Mol Microbiol. 2008 Apr;68(2):246-54. doi: 10.1111/j.1365-2958.2008.06147.x. Mol Microbiol. 2008. PMID: 18366437 Review.
-
Coupling of DNA replication and mitosis by fission yeast rad4/cut5.J Cell Sci Suppl. 1994;18:57-61. doi: 10.1242/jcs.1994.supplement_18.8. J Cell Sci Suppl. 1994. PMID: 7883793 Review.
Cited by
-
Inner nuclear membrane protein Lem2 facilitates Rad3-mediated checkpoint signaling under replication stress induced by nucleotide depletion in fission yeast.Cell Signal. 2016 Apr;28(4):235-45. doi: 10.1016/j.cellsig.2015.12.009. Epub 2015 Dec 31. Cell Signal. 2016. PMID: 26746798 Free PMC article.
-
Checkpoint functions of RecQ helicases at perturbed DNA replication fork.Curr Genet. 2021 Jun;67(3):369-382. doi: 10.1007/s00294-020-01147-y. Epub 2021 Jan 11. Curr Genet. 2021. PMID: 33427950 Review.
-
Hydroxyurea Induces Cytokinesis Arrest in Cells Expressing a Mutated Sterol-14α-Demethylase in the Ergosterol Biosynthesis Pathway.Genetics. 2016 Nov;204(3):959-973. doi: 10.1534/genetics.116.191536. Epub 2016 Aug 31. Genetics. 2016. PMID: 27585850 Free PMC article.
-
A tel2 Mutation That Destabilizes the Tel2-Tti1-Tti2 Complex Eliminates Rad3ATR Kinase Signaling in the DNA Replication Checkpoint and Leads to Telomere Shortening in Fission Yeast.Mol Cell Biol. 2019 Sep 27;39(20):e00175-19. doi: 10.1128/MCB.00175-19. Print 2019 Oct 15. Mol Cell Biol. 2019. PMID: 31332096 Free PMC article.
-
RecQ DNA Helicase Rqh1 Promotes Rad3ATR Kinase Signaling in the DNA Replication Checkpoint Pathway of Fission Yeast.Mol Cell Biol. 2020 Aug 14;40(17):e00145-20. doi: 10.1128/MCB.00145-20. Print 2020 Aug 14. Mol Cell Biol. 2020. PMID: 32541066 Free PMC article.
References
-
- Kelly TJ, Brown GW (2000) Regulation of chromosome replication. Annu Rev Biochem 69: 829–880. - PubMed
-
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M (2010) Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem 79: 89–130. - PubMed
-
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, et al. (2005) Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913. - PubMed
-
- Boddy MN, Furnari B, Mondesert O, Russell P (1998) Replication Checkpoint Enforced by Kinases Cds1 and Chk1. Science 280: 909–912. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

