The highly conserved MraZ protein is a transcriptional regulator in Escherichia coli
- PMID: 24659771
- PMCID: PMC4010979
- DOI: 10.1128/JB.01370-13
The highly conserved MraZ protein is a transcriptional regulator in Escherichia coli
Abstract
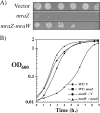
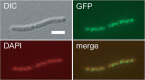
The mraZ and mraW genes are highly conserved in bacteria, both in sequence and in their position at the head of the division and cell wall (dcw) gene cluster. Located directly upstream of the mraZ gene, the Pmra promoter drives the transcription of mraZ and mraW, as well as many essential cell division and cell wall genes, but no regulator of Pmra has been found to date. Although MraZ has structural similarity to the AbrB transition state regulator and the MazE antitoxin and MraW is known to methylate the 16S rRNA, mraZ and mraW null mutants have no detectable phenotypes. Here we show that overproduction of Escherichia coli MraZ inhibited cell division and was lethal in rich medium at high induction levels and in minimal medium at low induction levels. Co-overproduction of MraW suppressed MraZ toxicity, and loss of MraW enhanced MraZ toxicity, suggesting that MraZ and MraW have antagonistic functions. MraZ-green fluorescent protein localized to the nucleoid, suggesting that it binds DNA. Consistent with this idea, purified MraZ directly bound a region of DNA containing three direct repeats between Pmra and the mraZ gene. Excess MraZ reduced the expression of an mraZ-lacZ reporter, suggesting that MraZ acts as a repressor of Pmra, whereas a DNA-binding mutant form of MraZ failed to repress expression. Transcriptome sequencing (RNA-seq) analysis suggested that MraZ also regulates the expression of genes outside the dcw cluster. In support of this, purified MraZ could directly bind to a putative operator site upstream of mioC, one of the repressed genes identified by RNA-seq.
Figures






Similar articles
-
mraW, an essential gene at the dcw cluster of Escherichia coli codes for a cytoplasmic protein with methyltransferase activity.Biochimie. 1999 Aug-Sep;81(8-9):879-88. doi: 10.1016/s0300-9084(99)00208-4. Biochimie. 1999. PMID: 10572301
-
MraZ Transcriptionally Controls the Critical Level of FtsL Required for Focusing Z-Rings and Kickstarting Septation in Bacillus subtilis.J Bacteriol. 2022 Sep 20;204(9):e0024322. doi: 10.1128/jb.00243-22. Epub 2022 Aug 9. J Bacteriol. 2022. PMID: 35943250 Free PMC article.
-
Contribution of the Pmra promoter to expression of genes in the Escherichia coli mra cluster of cell envelope biosynthesis and cell division genes.J Bacteriol. 1998 Sep;180(17):4406-12. doi: 10.1128/JB.180.17.4406-4412.1998. J Bacteriol. 1998. PMID: 9721276 Free PMC article.
-
Beyond a Ribosomal RNA Methyltransferase, the Wider Role of MraW in DNA Methylation, Motility and Colonization in Escherichia coli O157:H7.Front Microbiol. 2019 Nov 13;10:2520. doi: 10.3389/fmicb.2019.02520. eCollection 2019. Front Microbiol. 2019. PMID: 31798540 Free PMC article.
-
Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins.FEBS Lett. 2005 Oct 24;579(25):5669-74. doi: 10.1016/j.febslet.2005.09.045. Epub 2005 Oct 4. FEBS Lett. 2005. PMID: 16223496
Cited by
-
The Conserved Dcw Gene Cluster of R. sphaeroides Is Preceded by an Uncommonly Extended 5' Leader Featuring the sRNA UpsM.PLoS One. 2016 Nov 1;11(11):e0165694. doi: 10.1371/journal.pone.0165694. eCollection 2016. PLoS One. 2016. PMID: 27802301 Free PMC article.
-
New Global Insights on the Regulation of the Biphasic Life Cycle and Virulence Via ClpP-Dependent Proteolysis in Legionella pneumophila.Mol Cell Proteomics. 2022 May;21(5):100233. doi: 10.1016/j.mcpro.2022.100233. Epub 2022 Apr 12. Mol Cell Proteomics. 2022. PMID: 35427813 Free PMC article.
-
Reconstruction of Transcription Control Networks in Mollicutes by High-Throughput Identification of Promoters.Front Microbiol. 2016 Dec 6;7:1977. doi: 10.3389/fmicb.2016.01977. eCollection 2016. Front Microbiol. 2016. PMID: 27999573 Free PMC article.
-
Phylogenomics of expanding uncultured environmental Tenericutes provides insights into their pathogenicity and evolutionary relationship with Bacilli.BMC Genomics. 2020 Jun 17;21(1):408. doi: 10.1186/s12864-020-06807-4. BMC Genomics. 2020. PMID: 32552739 Free PMC article.
-
The Bacterial MtrAB Two-Component System Regulates the Cell Wall Homeostasis Responding to Environmental Alkaline Stress.Microbiol Spectr. 2022 Oct 26;10(5):e0231122. doi: 10.1128/spectrum.02311-22. Epub 2022 Sep 8. Microbiol Spectr. 2022. PMID: 36073914 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

