Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis
- PMID: 24655370
- PMCID: PMC4056528
- DOI: 10.1186/gb-2014-15-3-r52
Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis
Abstract
Background: Osteogenesis is a highly regulated developmental process and continues during the turnover and repair of mature bone. Runx2, the master regulator of osteoblastogenesis, directs a transcriptional program essential for bone formation through genetic and epigenetic mechanisms. While individual Runx2 gene targets have been identified, further insights into the broad spectrum of Runx2 functions required for osteogenesis are needed.
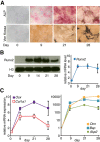
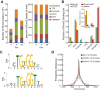
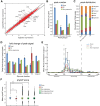
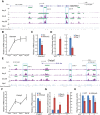
Results: By performing genome-wide characterization of Runx2 binding at the three major stages of osteoblast differentiation--proliferation, matrix deposition and mineralization--we identify Runx2-dependent regulatory networks driving bone formation. Using chromatin immunoprecipitation followed by high-throughput sequencing over the course of these stages, we identify approximately 80,000 significantly enriched regions of Runx2 binding throughout the mouse genome. These binding events exhibit distinct patterns during osteogenesis, and are associated with proximal promoters and also non-promoter regions: upstream, introns, exons, transcription termination site regions, and intergenic regions. These peaks were partitioned into clusters that are associated with genes in complex biological processes that support bone formation. Using Affymetrix expression profiling of differentiating osteoblasts depleted of Runx2, we identify novel Runx2 targets including Ezh2, a critical epigenetic regulator; Crabp2, a retinoic acid signaling component; Adamts4 and Tnfrsf19, two remodelers of the extracellular matrix. We demonstrate by luciferase assays that these novel biological targets are regulated by Runx2 occupancy at non-promoter regions.
Conclusions: Our data establish that Runx2 interactions with chromatin across the genome reveal novel genes, pathways and transcriptional mechanisms that contribute to the regulation of osteoblastogenesis.
Figures


Similar articles
-
Genome-wide DNase hypersensitivity, and occupancy of RUNX2 and CTCF reveal a highly dynamic gene regulome during MC3T3 pre-osteoblast differentiation.PLoS One. 2017 Nov 27;12(11):e0188056. doi: 10.1371/journal.pone.0188056. eCollection 2017. PLoS One. 2017. PMID: 29176792 Free PMC article.
-
Comparative analysis of osteoblast gene expression profiles and Runx2 genomic occupancy of mouse and human osteoblasts in vitro.Gene. 2017 Aug 30;626:119-131. doi: 10.1016/j.gene.2017.05.028. Epub 2017 May 11. Gene. 2017. PMID: 28502869
-
Expression of the ectodomain-releasing protease ADAM17 is directly regulated by the osteosarcoma and bone-related transcription factor RUNX2.J Cell Biochem. 2018 Nov;119(10):8204-8219. doi: 10.1002/jcb.26832. Epub 2018 Jun 19. J Cell Biochem. 2018. PMID: 29923217 Free PMC article.
-
Networks and hubs for the transcriptional control of osteoblastogenesis.Rev Endocr Metab Disord. 2006 Jun;7(1-2):1-16. doi: 10.1007/s11154-006-9001-5. Rev Endocr Metab Disord. 2006. PMID: 17051438 Review.
-
Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors.Crit Rev Eukaryot Gene Expr. 2004;14(1-2):1-41. Crit Rev Eukaryot Gene Expr. 2004. PMID: 15104525 Review.
Cited by
-
Runx2+ Niche Cells Maintain Incisor Mesenchymal Tissue Homeostasis through IGF Signaling.Cell Rep. 2020 Aug 11;32(6):108007. doi: 10.1016/j.celrep.2020.108007. Cell Rep. 2020. PMID: 32783935 Free PMC article.
-
Mll-COMPASS complexes mediate H3K4me3 enrichment and transcription of the osteoblast master gene Runx2/p57 in osteoblasts.J Cell Physiol. 2019 May;234(5):6244-6253. doi: 10.1002/jcp.27355. Epub 2018 Sep 7. J Cell Physiol. 2019. PMID: 30256410 Free PMC article.
-
Emerging RUNX2-Mediated Gene Regulatory Mechanisms Consisting of Multi-Layered Regulatory Networks in Skeletal Development.Int J Mol Sci. 2023 Feb 3;24(3):2979. doi: 10.3390/ijms24032979. Int J Mol Sci. 2023. PMID: 36769300 Free PMC article. Review.
-
WWOX and p53 Dysregulation Synergize to Drive the Development of Osteosarcoma.Cancer Res. 2016 Oct 15;76(20):6107-6117. doi: 10.1158/0008-5472.CAN-16-0621. Epub 2016 Aug 22. Cancer Res. 2016. PMID: 27550453 Free PMC article.
-
A unique stylopod patterning mechanism by Shox2-controlled osteogenesis.Development. 2016 Jul 15;143(14):2548-60. doi: 10.1242/dev.138750. Epub 2016 Jun 10. Development. 2016. PMID: 27287812 Free PMC article.
References
-
- Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:1–16. - PubMed
-
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2012;13:27–38. - PubMed
-
- Hecht J, Seitz V, Urban M, Wagner F, Robinson PN, Stiege A, Dieterich C, Kornak U, Wilkening U, Brieske N, Zwingman C, Kidess A, Stricker S, Mundlos S. Detection of novel skeletogenesis target genes by comprehensive analysis of a Runx2(-/-) mouse model. Gene Expr Patterns. 2007;7:102–112. doi: 10.1016/j.modgep.2006.05.014. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

