Identification of binding sites in Huntingtin for the Huntingtin Interacting Proteins HIP14 and HIP14L
- PMID: 24651384
- PMCID: PMC3947954
- DOI: 10.1371/journal.pone.0090669
Identification of binding sites in Huntingtin for the Huntingtin Interacting Proteins HIP14 and HIP14L
Abstract
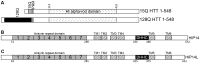
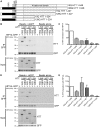
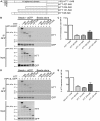
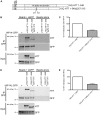
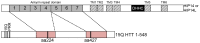
Huntington disease is an adult onset neurodegenerative disease characterized by motor, cognitive, and psychiatric dysfunction, caused by a CAG expansion in the HTT gene. Huntingtin Interacting Protein 14 (HIP14) and Huntingtin Interacting Protein 14-like (HIP14L) are palmitoyl acyltransferases (PATs), enzymes that mediate the post-translational addition of long chain fatty acids to proteins in a process called palmitoylation. HIP14 and HIP14L interact with and palmitoylate HTT and are unique among PATs as they are the only two that have an ankyrin repeat domain, which mediates the interaction between HIP14 and HTT. These enzymes show reduced interaction with and palmitoylation of mutant HTT, leading to increased mutant HTT inclusion formation and toxicity. The interaction between HIP14 and HTT goes beyond that of only an enzyme-substrate interaction as HTT is essential for the full enzymatic activity of HIP14. It is important to further understand and characterize the interactions of HTT with HIP14 and HIP14L to guide future efforts to target and enhance this interaction and increase enzyme activity to remediate palmitoylation of HTT and their substrates, as well as to understand the relationship between the three proteins. HIP14 and HIP14L have been previously shown to interact with HTT amino acids 1-548. Here the interaction of HIP14 and HIP14L with N- and C-terminal HTT 1-548 deletion mutations was assessed. We show that HTT amino acids 1-548 were sufficient for full interaction of HTT with HIP14 and HIP14L, but partial interaction was also possible with HTT 1-427 and HTT 224-548. To further characterize the binding domain we assessed the interaction of HIP14-GFP and HIP14L-GFP with 15Q HTT 1-548Δ257-315. Both enzymes showed reduced but not abolished interaction with 15Q HTT 1-548Δ257-315. This suggests that two potential binding domains exist, one around residues 224 and the other around 427, for the PAT enzymes HIP14 and HIP14L.
Conflict of interest statement
Figures
Similar articles
-
Hip14l-deficient mice develop neuropathological and behavioural features of Huntington disease.Hum Mol Genet. 2013 Feb 1;22(3):452-65. doi: 10.1093/hmg/dds441. Epub 2012 Oct 16. Hum Mol Genet. 2013. PMID: 23077216
-
Aberrant palmitoylation in Huntington disease.Biochem Soc Trans. 2015 Apr;43(2):205-10. doi: 10.1042/BST20140242. Biochem Soc Trans. 2015. PMID: 25849918 Review.
-
Huntingtin interacting proteins 14 and 14-like are required for chorioallantoic fusion during early placental development.Dev Biol. 2015 Jan 15;397(2):257-66. doi: 10.1016/j.ydbio.2014.11.018. Epub 2014 Dec 3. Dev Biol. 2015. PMID: 25478910
-
Wild-type HTT modulates the enzymatic activity of the neuronal palmitoyl transferase HIP14.Hum Mol Genet. 2011 Sep 1;20(17):3356-65. doi: 10.1093/hmg/ddr242. Epub 2011 Jun 2. Hum Mol Genet. 2011. PMID: 21636527 Free PMC article.
-
Putting proteins in their place: palmitoylation in Huntington disease and other neuropsychiatric diseases.Prog Neurobiol. 2012 May;97(2):220-38. doi: 10.1016/j.pneurobio.2011.11.002. Epub 2011 Dec 7. Prog Neurobiol. 2012. PMID: 22155432 Review.
Cited by
-
Altered Regulation of Striatal Neuronal N-Methyl-D-Aspartate Receptor Trafficking by Palmitoylation in Huntington Disease Mouse Model.Front Synaptic Neurosci. 2019 Feb 21;11:3. doi: 10.3389/fnsyn.2019.00003. eCollection 2019. Front Synaptic Neurosci. 2019. PMID: 30846936 Free PMC article.
-
Characterization of huntingtin interactomes and their dynamic responses in living cells by proximity proteomics.J Neurochem. 2023 Feb;164(4):512-528. doi: 10.1111/jnc.15726. Epub 2022 Nov 27. J Neurochem. 2023. PMID: 36437609 Free PMC article.
-
Palmitoylation of ULK1 by ZDHHC13 plays a crucial role in autophagy.Nat Commun. 2024 Aug 21;15(1):7194. doi: 10.1038/s41467-024-51402-w. Nat Commun. 2024. PMID: 39169022 Free PMC article.
-
Mutant Huntingtin Protein Interaction Map Implicates Dysregulation of Multiple Cellular Pathways in Neurodegeneration of Huntington's Disease.J Huntingtons Dis. 2022;11(3):243-267. doi: 10.3233/JHD-220538. J Huntingtons Dis. 2022. PMID: 35871359 Free PMC article.
-
Structural Basis for Substrate Recognition by the Ankyrin Repeat Domain of Human DHHC17 Palmitoyltransferase.Structure. 2017 Sep 5;25(9):1337-1347.e6. doi: 10.1016/j.str.2017.06.018. Epub 2017 Jul 27. Structure. 2017. PMID: 28757145 Free PMC article.
References
-
- The Huntington's disease collaborative research group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72: 971–983. - PubMed
-
- Kalchman MA, Graham RK, Xia G, Koide HB, Hodgson JG, et al. (1996) Huntingtin is ubiquitinated and interacts with a specific ubiquitin-conjugating enzyme. J Biol Chem 271: 19385–19394. - PubMed
-
- Singaraja RR, Hadano S, Metzler M, Givan S, Wellington CL, et al. (2002) HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Human Molecular Genetics 11: 2815–2828. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

