Components of Golgi-to-vacuole trafficking are required for nitrogen- and TORC1-responsive regulation of the yeast GATA factors
- PMID: 24644271
- PMCID: PMC4082702
- DOI: 10.1002/mbo3.168
Components of Golgi-to-vacuole trafficking are required for nitrogen- and TORC1-responsive regulation of the yeast GATA factors
Abstract
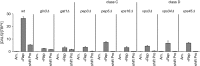
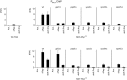
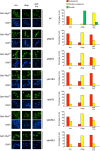
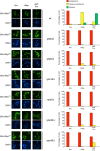
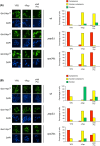
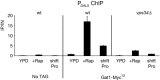
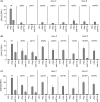
Nitrogen catabolite repression (NCR) is the regulatory pathway through which Saccharomyces cerevisiae responds to the available nitrogen status and selectively utilizes rich nitrogen sources in preference to poor ones. Expression of NCR-sensitive genes is mediated by two transcription activators, Gln3 and Gat1, in response to provision of a poorly used nitrogen source or following treatment with the TORC1 inhibitor, rapamycin. During nitrogen excess, the transcription activators are sequestered in the cytoplasm in a Ure2-dependent fashion. Here, we show that Vps components are required for Gln3 localization and function in response to rapamycin treatment when cells are grown in defined yeast nitrogen base but not in complex yeast peptone dextrose medium. On the other hand, Gat1 function was altered in vps mutants in all conditions tested. A significant fraction of Gat1, like Gln3, is associated with light intracellular membranes. Further, our results are consistent with the possibility that Ure2 might function downstream of the Vps components during the control of GATA factor-mediated gene expression. These observations demonstrate distinct media-dependent requirements of vesicular trafficking components for wild-type responses of GATA factor localization and function. As a result, the current model describing participation of Vps system components in events associated with translocation of Gln3 into the nucleus following rapamycin treatment or growth in nitrogen-poor medium requires modification.
Keywords: GATA factor; nitrogen availability; rapamycin; vesicular trafficking; yeast.
© 2014 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures

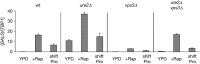
Similar articles
-
Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae.J Biol Chem. 2010 Jun 4;285(23):17880-95. doi: 10.1074/jbc.M109.085712. Epub 2010 Apr 8. J Biol Chem. 2010. PMID: 20378536 Free PMC article.
-
Nitrogen starvation and TorC1 inhibition differentially affect nuclear localization of the Gln3 and Gat1 transcription factors through the rare glutamine tRNACUG in Saccharomyces cerevisiae.Genetics. 2015 Feb;199(2):455-74. doi: 10.1534/genetics.114.173831. Epub 2014 Dec 19. Genetics. 2015. PMID: 25527290 Free PMC article.
-
General Amino Acid Control and 14-3-3 Proteins Bmh1/2 Are Required for Nitrogen Catabolite Repression-Sensitive Regulation of Gln3 and Gat1 Localization.Genetics. 2017 Feb;205(2):633-655. doi: 10.1534/genetics.116.195800. Epub 2016 Dec 22. Genetics. 2017. PMID: 28007891 Free PMC article.
-
Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots.FEMS Microbiol Rev. 2002 Aug;26(3):223-38. doi: 10.1111/j.1574-6976.2002.tb00612.x. FEMS Microbiol Rev. 2002. PMID: 12165425 Free PMC article. Review.
-
Nitrogen catabolite repression in Saccharomyces cerevisiae.Mol Biotechnol. 1999 Aug;12(1):35-73. doi: 10.1385/MB:12:1:35. Mol Biotechnol. 1999. PMID: 10554772 Review.
Cited by
-
Vesicular Trafficking Systems Impact TORC1-Controlled Transcriptional Programs in Saccharomyces cerevisiae.G3 (Bethesda). 2016 Jan 6;6(3):641-52. doi: 10.1534/g3.115.023911. G3 (Bethesda). 2016. PMID: 26739646 Free PMC article.
-
Chromatin Regulators Ahc1p and Eaf3p Positively Influence Nitrogen Metabolism in Saccharomyces cerevisiae.Front Microbiol. 2022 May 10;13:883934. doi: 10.3389/fmicb.2022.883934. eCollection 2022. Front Microbiol. 2022. PMID: 35620110 Free PMC article.
-
GATA Factor Regulation in Excess Nitrogen Occurs Independently of Gtr-Ego Complex-Dependent TorC1 Activation.G3 (Bethesda). 2015 May 29;5(8):1625-38. doi: 10.1534/g3.115.019307. G3 (Bethesda). 2015. PMID: 26024867 Free PMC article.
-
The pleiotropic effects of the glutamate dehydrogenase (GDH) pathway in Saccharomyces cerevisiae.Microb Cell Fact. 2018 Nov 1;17(1):170. doi: 10.1186/s12934-018-1018-4. Microb Cell Fact. 2018. PMID: 30384856 Free PMC article. Review.
-
The GATA Transcription Factor Gaf1 Represses tRNAs, Inhibits Growth, and Extends Chronological Lifespan Downstream of Fission Yeast TORC1.Cell Rep. 2020 Mar 10;30(10):3240-3249.e4. doi: 10.1016/j.celrep.2020.02.058. Cell Rep. 2020. PMID: 32160533 Free PMC article.
References
-
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem. J. 2008;410:1–17. - PubMed
-
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. - PubMed
-
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 2009;35:563–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

