Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins
- PMID: 24637324
- PMCID: PMC3998805
- DOI: 10.1083/jcb.201305148
Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins
Abstract
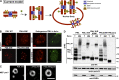
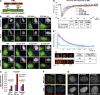
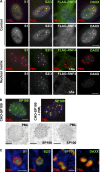
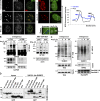
The promyelocytic leukemia (PML) protein organizes PML nuclear bodies (NBs), which are stress-responsive domains where many partner proteins accumulate. Here, we clarify the basis for NB formation and identify stress-induced partner sumoylation as the primary NB function. NB nucleation does not rely primarily on intermolecular interactions between the PML SUMO-interacting motif (SIM) and SUMO, but instead results from oxidation-mediated PML multimerization. Oxidized PML spherical meshes recruit UBC9, which enhances PML sumoylation, allow partner recruitment through SIM interactions, and ultimately enhance partner sumoylation. Intermolecular SUMO-SIM interactions then enforce partner sequestration within the NB inner core. Accordingly, oxidative stress enhances NB formation and global sumoylation in vivo. Some NB-associated sumoylated partners also become polyubiquitinated by RNF4, precipitating their proteasomal degradation. As several partners are protein-modifying enzymes, NBs could act as sensors that facilitate and confer oxidative stress sensitivity not only to sumoylation but also to other post-translational modifications, thereby explaining alterations of stress response upon PML or NB loss.
Figures
Similar articles
-
Requirement of PML SUMO interacting motif for RNF4- or arsenic trioxide-induced degradation of nuclear PML isoforms.PLoS One. 2012;7(9):e44949. doi: 10.1371/journal.pone.0044949. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23028697 Free PMC article.
-
SUMOylation promotes PML degradation during encephalomyocarditis virus infection.J Virol. 2010 Nov;84(22):11634-45. doi: 10.1128/JVI.01321-10. Epub 2010 Sep 8. J Virol. 2010. PMID: 20826694 Free PMC article.
-
SUMOylation regulates the number and size of promyelocytic leukemia-nuclear bodies (PML-NBs) and arsenic perturbs SUMO dynamics on PML by insolubilizing PML in THP-1 cells.Arch Toxicol. 2022 Feb;96(2):545-558. doi: 10.1007/s00204-021-03195-w. Epub 2022 Jan 10. Arch Toxicol. 2022. PMID: 35001170
-
A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics.Int J Biol Sci. 2010 Jan 12;6(1):51-67. doi: 10.7150/ijbs.6.51. Int J Biol Sci. 2010. PMID: 20087442 Free PMC article. Review.
-
PML nuclear bodies: assembly and oxidative stress-sensitive sumoylation.Nucleus. 2014;5(6):499-507. doi: 10.4161/19491034.2014.970104. Nucleus. 2014. PMID: 25482067 Free PMC article. Review.
Cited by
-
The pathophysiology of neurodegenerative disease: Disturbing the balance between phase separation and irreversible aggregation.Prog Mol Biol Transl Sci. 2020;174:187-223. doi: 10.1016/bs.pmbts.2020.04.021. Epub 2020 May 12. Prog Mol Biol Transl Sci. 2020. PMID: 32828466 Free PMC article. Review.
-
Pathogenic Mutations in the Valosin-containing Protein/p97(VCP) N-domain Inhibit the SUMOylation of VCP and Lead to Impaired Stress Response.J Biol Chem. 2016 Jul 1;291(27):14373-14384. doi: 10.1074/jbc.M116.729343. Epub 2016 May 13. J Biol Chem. 2016. PMID: 27226613 Free PMC article.
-
Nuclear body phase separation drives telomere clustering in ALT cancer cells.Mol Biol Cell. 2020 Aug 15;31(18):2048-2056. doi: 10.1091/mbc.E19-10-0589. Epub 2020 Jun 24. Mol Biol Cell. 2020. PMID: 32579423 Free PMC article.
-
Viruses in the Nucleus.Cold Spring Harb Perspect Biol. 2021 Aug 2;13(8):a039446. doi: 10.1101/cshperspect.a039446. Cold Spring Harb Perspect Biol. 2021. PMID: 33753405 Free PMC article. Review.
-
HIF-1α SUMOylation affects the stability and transcriptional activity of HIF-1α in human lens epithelial cells.Graefes Arch Clin Exp Ophthalmol. 2015 Aug;253(8):1279-90. doi: 10.1007/s00417-015-2999-x. Epub 2015 Apr 16. Graefes Arch Clin Exp Ophthalmol. 2015. PMID: 25877955
References
-
- Chiantore M.V., Vannucchi S., Accardi R., Tommasino M., Percario Z.A., Vaccari G., Affabris E., Fiorucci G., Romeo G. 2012. Interferon-β induces cellular senescence in cutaneous human papilloma virus-transformed human keratinocytes by affecting p53 transactivating activity. PLoS ONE. 7:e36909 10.1371/journal.pone.0036909 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

