Hydrogen peroxide sensing, signaling and regulation of transcription factors
- PMID: 24634836
- PMCID: PMC3953959
- DOI: 10.1016/j.redox.2014.02.006
Hydrogen peroxide sensing, signaling and regulation of transcription factors
Abstract
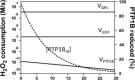
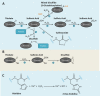
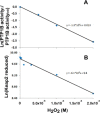
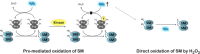
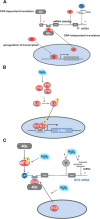
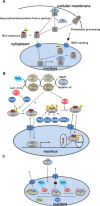
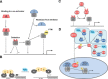
The regulatory mechanisms by which hydrogen peroxide (H2O2) modulates the activity of transcription factors in bacteria (OxyR and PerR), lower eukaryotes (Yap1, Maf1, Hsf1 and Msn2/4) and mammalian cells (AP-1, NRF2, CREB, HSF1, HIF-1, TP53, NF-κB, NOTCH, SP1 and SCREB-1) are reviewed. The complexity of regulatory networks increases throughout the phylogenetic tree, reaching a high level of complexity in mammalians. Multiple H2O2 sensors and pathways are triggered converging in the regulation of transcription factors at several levels: (1) synthesis of the transcription factor by upregulating transcription or increasing both mRNA stability and translation; (ii) stability of the transcription factor by decreasing its association with the ubiquitin E3 ligase complex or by inhibiting this complex; (iii) cytoplasm-nuclear traffic by exposing/masking nuclear localization signals, or by releasing the transcription factor from partners or from membrane anchors; and (iv) DNA binding and nuclear transactivation by modulating transcription factor affinity towards DNA, co-activators or repressors, and by targeting specific regions of chromatin to activate individual genes. We also discuss how H2O2 biological specificity results from diverse thiol protein sensors, with different reactivity of their sulfhydryl groups towards H2O2, being activated by different concentrations and times of exposure to H2O2. The specific regulation of local H2O2 concentrations is also crucial and results from H2O2 localized production and removal controlled by signals. Finally, we formulate equations to extract from typical experiments quantitative data concerning H2O2 reactivity with sensor molecules. Rate constants of 140 M(-1) s(-1) and ≥1.3 × 10(3) M(-1) s(-1) were estimated, respectively, for the reaction of H2O2 with KEAP1 and with an unknown target that mediates NRF2 protein synthesis. In conclusion, the multitude of H2O2 targets and mechanisms provides an opportunity for highly specific effects on gene regulation that depend on the cell type and on signals received from the cellular microenvironment.
Keywords: AD, activation domain; Cytosol-nuclear traffic; DNA binding and transactivation; ER, endoplasmic reticulum; GPx, glutathione peroxidases; Localized H2O2 concentrations; NES, nuclear exporting signal; NLS, nuclear localization signal; PHD, prolyl hydroxylase; Prxs, peroxiredoxins; Rate constants; Redox signaling; TF, transcription factor; Thiol reactivity; Ub, Ubiquitin.
Figures


Similar articles
-
OxyR: a molecular code for redox sensing?Sci STKE. 2002 Nov 5;2002(157):pe46. doi: 10.1126/stke.2002.157.pe46. Sci STKE. 2002. PMID: 12419849 Review.
-
OxyR2 Functions as a Three-state Redox Switch to Tightly Regulate Production of Prx2, a Peroxiredoxin of Vibrio vulnificus.J Biol Chem. 2016 Jul 29;291(31):16038-47. doi: 10.1074/jbc.M115.710343. Epub 2016 Jun 6. J Biol Chem. 2016. PMID: 27268058 Free PMC article.
-
The Roles of Peroxiredoxin and Thioredoxin in Hydrogen Peroxide Sensing and in Signal Transduction.Mol Cells. 2016 Jan;39(1):65-71. doi: 10.14348/molcells.2016.2349. Epub 2016 Jan 25. Mol Cells. 2016. PMID: 26813662 Free PMC article. Review.
-
Thiol-based H2O2 signalling in microbial systems.Redox Biol. 2014 Feb 3;2:395-9. doi: 10.1016/j.redox.2014.01.015. eCollection 2014. Redox Biol. 2014. PMID: 24563858 Free PMC article. Review.
-
Peroxiredoxins and sulfiredoxin at the crossroads of the NO and H2O2 signaling pathways.Methods Enzymol. 2013;527:113-28. doi: 10.1016/B978-0-12-405882-8.00006-4. Methods Enzymol. 2013. PMID: 23830628
Cited by
-
Long-term enzyme replacement therapy in Fabry patients protects against oxidative and inflammatory process.Naunyn Schmiedebergs Arch Pharmacol. 2024 Oct 22. doi: 10.1007/s00210-024-03499-5. Online ahead of print. Naunyn Schmiedebergs Arch Pharmacol. 2024. PMID: 39436428
-
Light and hydrogen peroxide inhibit C. elegans Feeding through gustatory receptor orthologs and pharyngeal neurons.Neuron. 2015 Feb 18;85(4):804-18. doi: 10.1016/j.neuron.2014.12.061. Epub 2015 Jan 29. Neuron. 2015. PMID: 25640076 Free PMC article.
-
In Vitro Evaluation of Antidiabetic and Anti-Inflammatory Activities of Polyphenolic-Rich Extracts from Anchusa officinalis and Melilotus officinalis.ACS Omega. 2020 May 22;5(22):13014-13022. doi: 10.1021/acsomega.0c00929. eCollection 2020 Jun 9. ACS Omega. 2020. PMID: 32548486 Free PMC article.
-
Emerging trends and hotspots of Nuclear factor erythroid 2-related factor 2 in nervous system diseases.World J Clin Cases. 2023 Nov 16;11(32):7833-7851. doi: 10.12998/wjcc.v11.i32.7833. World J Clin Cases. 2023. PMID: 38073678 Free PMC article.
-
The dynamic interplay between cardiac mitochondrial health and myocardial structural remodeling in metabolic heart disease, aging, and heart failure.J Cardiovasc Aging. 2023 Jan;3(1):9. doi: 10.20517/jca.2022.42. Epub 2023 Jan 3. J Cardiovasc Aging. 2023. PMID: 36742465 Free PMC article.
References
-
- Czech MP, Lawrence JC, Lynn WS. Evidence for the involvement of sulfhydryl oxidation in the regulation of fat cell hexose transport by insulin. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:4173–4177. doi: 10.1073/pnas.71.10.4173. 4372610 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

