Metabolic versatility in Haemophilus influenzae: a metabolomic and genomic analysis
- PMID: 24624122
- PMCID: PMC3941224
- DOI: 10.3389/fmicb.2014.00069
Metabolic versatility in Haemophilus influenzae: a metabolomic and genomic analysis
Abstract
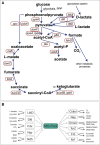
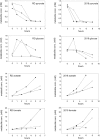
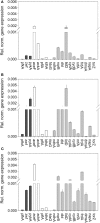
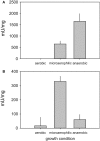
Haemophilus influenzae is a host adapted human pathogen known to contribute to a variety of acute and chronic diseases of the upper and lower respiratory tract as well as the middle ear. At the sites of infection as well as during growth as a commensal the environmental conditions encountered by H. influenzae will vary significantly, especially in terms of oxygen availability, however, the mechanisms by which the bacteria can adapt their metabolism to cope with such changes have not been studied in detail. Using targeted metabolomics the spectrum of metabolites produced during growth of H. influenzae on glucose in RPMI-based medium was found to change from acetate as the main product during aerobic growth to formate as the major product during anaerobic growth. This change in end-product is likely caused by a switch in the major route of pyruvate degradation. Neither lactate nor succinate or fumarate were major products of H. influenzae growth under any condition studied. Gene expression studies and enzyme activity data revealed that despite an identical genetic makeup and very similar metabolite production profiles, H. influenzae strain Rd appeared to favor glucose degradation via the pentose phosphate pathway, while strain 2019, a clinical isolate, showed higher expression of enzymes involved in glycolysis. Components of the respiratory chain were most highly expressed during microaerophilic and anaerobic growth in both strains, but again clear differences existed in the expression of genes associated e.g., with NADH oxidation, nitrate and nitrite reduction in the two strains studied. Together our results indicate that H. influenzae uses a specialized type of metabolism that could be termed "respiration assisted fermentation" where the respiratory chain likely serves to alleviate redox imbalances caused by incomplete glucose oxidation, and at the same time provides a means of converting a variety of compounds including nitrite and nitrate that arise as part of the host defence mechanisms.
Keywords: Haemophilus influenzae; carbon metabolism; enzymes; gene expression; proton NMR.
Figures
Similar articles
-
Haemophilus influenzae Glucose Catabolism Leading to Production of the Immunometabolite Acetate Has a Key Contribution to the Host Airway-Pathogen Interplay.ACS Infect Dis. 2020 Mar 13;6(3):406-421. doi: 10.1021/acsinfecdis.9b00359. Epub 2020 Feb 10. ACS Infect Dis. 2020. PMID: 31933358
-
Comprehensive Proteomic and Metabolomic Signatures of Nontypeable Haemophilus influenzae-Induced Acute Otitis Media Reveal Bacterial Aerobic Respiration in an Immunosuppressed Environment.Mol Cell Proteomics. 2016 Mar;15(3):1117-38. doi: 10.1074/mcp.M115.052498. Epub 2015 Dec 28. Mol Cell Proteomics. 2016. PMID: 26711468 Free PMC article.
-
In vivo gene expression profile of Haemophilus influenzae during human pneumonia.Microbiol Spectr. 2023 Sep 14;11(5):e0163923. doi: 10.1128/spectrum.01639-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37707456 Free PMC article.
-
Glucose and lactate metabolism by Actinomyces naeslundii.Crit Rev Oral Biol Med. 1999;10(4):487-503. doi: 10.1177/10454411990100040501. Crit Rev Oral Biol Med. 1999. PMID: 10634585 Review.
-
Haemophilus influenzae and oxidative stress.Front Cell Infect Microbiol. 2012 Mar 28;2:40. doi: 10.3389/fcimb.2012.00040. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919631 Free PMC article. Review.
Cited by
-
The DmsABC S-oxide reductase is an essential component of a novel, hypochlorite-inducible system of extracellular stress defense in Haemophilus influenzae.Front Microbiol. 2024 Apr 4;15:1359513. doi: 10.3389/fmicb.2024.1359513. eCollection 2024. Front Microbiol. 2024. PMID: 38638903 Free PMC article.
-
A Novel, Molybdenum-Containing Methionine Sulfoxide Reductase Supports Survival of Haemophilus influenzae in an In vivo Model of Infection.Front Microbiol. 2016 Nov 14;7:1743. doi: 10.3389/fmicb.2016.01743. eCollection 2016. Front Microbiol. 2016. PMID: 27933034 Free PMC article.
-
The DmsABC Sulfoxide Reductase Supports Virulence in Non-typeable Haemophilus influenzae.Front Microbiol. 2021 Jul 22;12:686833. doi: 10.3389/fmicb.2021.686833. eCollection 2021. Front Microbiol. 2021. PMID: 34367088 Free PMC article.
-
Total sputum nitrate/nitrite is associated with exacerbations and Pseudomonas aeruginosa colonisation in bronchiectasis.ERJ Open Res. 2024 Jul 22;10(4):01045-2023. doi: 10.1183/23120541.01045-2023. eCollection 2024 Jul. ERJ Open Res. 2024. PMID: 39040581 Free PMC article.
-
The Alternative Sigma Factor RpoE2 Is Involved in the Stress Response to Hypochlorite and in vivo Survival of Haemophilus influenzae.Front Microbiol. 2021 Feb 12;12:637213. doi: 10.3389/fmicb.2021.637213. eCollection 2021. Front Microbiol. 2021. PMID: 33643271 Free PMC article.
References
-
- Alexander H. E. (1965). “The haemophilus group,” in Bacterial and Mycotic Infections of Man, eds Dabos R. J., Hirsch J. G. (London: Pitman Medical Publishing Co. Ltd; ).
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., et al. (2005). “Current protocols in molecular biology,” in Current Protocols in Molecular Biology, ed Janssen K. (Hoboken, NJ: John Wiley & Sons Inc; ).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

