Virus-induced transcriptional changes in the brain include the differential expression of genes associated with interferon, apoptosis, interleukin 17 receptor A, and glutamate signaling as well as flavivirus-specific upregulation of tRNA synthetases
- PMID: 24618253
- PMCID: PMC3952157
- DOI: 10.1128/mBio.00902-14
Virus-induced transcriptional changes in the brain include the differential expression of genes associated with interferon, apoptosis, interleukin 17 receptor A, and glutamate signaling as well as flavivirus-specific upregulation of tRNA synthetases
Abstract
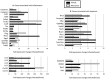
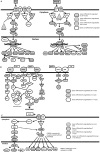
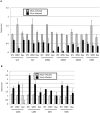
Flaviviruses, particularly Japanese encephalitis virus (JEV) and West Nile virus (WNV), are important causes of virus-induced central nervous system (CNS) disease in humans. We used microarray analysis to identify cellular genes that are differentially regulated following infection of the brain with JEV (P3) or WNV (New York 99). Gene expression data for these flaviviruses were compared to those obtained following infection of the brain with reovirus (type 3 Dearing), an unrelated neurotropic virus. We found that a large number of genes were up-regulated by all three viruses (using the criteria of a change of >2-fold and a P value of <0.001), including genes associated with interferon signaling, the immune system, inflammation, and cell death/survival signaling. In addition, genes associated with glutamate signaling were down-regulated in infections with all three viruses (criteria, a >2-fold change and a P value of <0.001). These genes may serve as broad-spectrum therapeutic targets for virus-induced CNS disease. A distinct set of genes were up-regulated following flavivirus infection but not following infection with reovirus. These genes were associated with tRNA charging and may serve as therapeutic targets for flavivirus-induced CNS disease. IMPORTANCE Viral infections of the central nervous system (CNS) are an important cause of morbidity and mortality. Treatment options for virus-induced CNS disease are limited, and for many clinically important neurotropic viruses, no specific therapy of proven benefit is currently available. We performed microarray analysis to identify genes that are differentially regulated in the brain following virus infection in order to identify pathways that might provide novel therapeutic targets for virus-induced CNS disease. Although several studies have described gene expression changes following virus infection of the brain, this report is the first to directly compare large-scale gene expression data from different viruses. We identified genes that are differentially regulated in infection of the brain with viruses from different families and those which appear to be specific to flavivirus infections.
Figures


Similar articles
-
A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection.J Virol. 2017 Mar 29;91(8):e02388-16. doi: 10.1128/JVI.02388-16. Print 2017 Apr 15. J Virol. 2017. PMID: 28148804 Free PMC article.
-
The critical role of interleukin-6 in protection against neurotropic flavivirus infection.Front Cell Infect Microbiol. 2023 Nov 16;13:1275823. doi: 10.3389/fcimb.2023.1275823. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38053527 Free PMC article.
-
Survival of mice immunized with monoclonal antibodies against glycoprotein E of Japanese encephalitis virus before or after infection with Japanese encephalitis, West Nile, and Dengue viruses.Acta Virol. 2008;52(4):219-24. Acta Virol. 2008. PMID: 19143477
-
Regulation of Apoptosis during Flavivirus Infection.Viruses. 2017 Aug 28;9(9):243. doi: 10.3390/v9090243. Viruses. 2017. PMID: 28846635 Free PMC article. Review.
-
Astrocytes in Flavivirus Infections.Int J Mol Sci. 2019 Feb 6;20(3):691. doi: 10.3390/ijms20030691. Int J Mol Sci. 2019. PMID: 30736273 Free PMC article. Review.
Cited by
-
The flavivirus dengue induces hypertrophy of white matter astrocytes.J Neurovirol. 2016 Dec;22(6):831-839. doi: 10.1007/s13365-016-0461-4. Epub 2016 Jun 6. J Neurovirol. 2016. PMID: 27273075 Free PMC article.
-
High throughput proteomic analysis and a comparative review identify the nuclear chaperone, Nucleophosmin among the common set of proteins modulated in Chikungunya virus infection.J Proteomics. 2015 Apr 29;120:126-41. doi: 10.1016/j.jprot.2015.03.007. Epub 2015 Mar 14. J Proteomics. 2015. PMID: 25782748 Free PMC article. Review.
-
ISG12a Restricts Hepatitis C Virus Infection through the Ubiquitination-Dependent Degradation Pathway.J Virol. 2016 Jul 11;90(15):6832-45. doi: 10.1128/JVI.00352-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27194766 Free PMC article.
-
Depletion of Microglia in an Ex Vivo Brain Slice Culture Model of West Nile Virus Infection Leads to Increased Viral Titers and Cell Death.Microbiol Spectr. 2022 Apr 27;10(2):e0068522. doi: 10.1128/spectrum.00685-22. Epub 2022 Apr 12. Microbiol Spectr. 2022. PMID: 35412380 Free PMC article.
-
Autoimmune encephalitis and its relation to infection.Curr Neurol Neurosci Rep. 2015 Mar;15(3):3. doi: 10.1007/s11910-015-0529-1. Curr Neurol Neurosci Rep. 2015. PMID: 25637289 Review.
References
-
- German AC, Myint KS, Mai NT, Pomeroy I, Phu NH, Tzartos J, Winter P, Collett J, Farrar J, Barrett A, Kipar A, Esiri MM, Solomon T. 2006. A preliminary neuropathological study of Japanese encephalitis in humans and a mouse model. Trans. R. Soc. Trop. Med. Hyg. 100:1135–1145. 10.1016/j.trstmh.2006.02.008 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
