Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation
- PMID: 24613413
- PMCID: PMC3992437
- DOI: 10.1016/j.ccr.2014.01.032
Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation
Abstract
Clonal evolution and intratumoral heterogeneity drive cancer progression through unknown molecular mechanisms. To address this issue, functional differences between single T cell acute lymphoblastic leukemia (T-ALL) clones were assessed using a zebrafish transgenic model. Functional variation was observed within individual clones, with a minority of clones enhancing growth rate and leukemia-propagating potential with time. Akt pathway activation was acquired in a subset of these evolved clones, which increased the number of leukemia-propagating cells through activating mTORC1, elevated growth rate likely by stabilizing the Myc protein, and rendered cells resistant to dexamethasone, which was reversed by combined treatment with an Akt inhibitor. Thus, T-ALL clones spontaneously and continuously evolve to drive leukemia progression even in the absence of therapy-induced selection.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

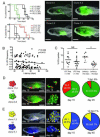
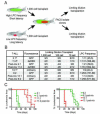
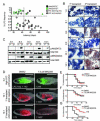
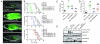
Comment in
-
Leukemia propagating cells Akt up.Cancer Cell. 2014 Mar 17;25(3):263-5. doi: 10.1016/j.ccr.2014.02.022. Cancer Cell. 2014. PMID: 24651006
Similar articles
-
Leukemia propagating cells Akt up.Cancer Cell. 2014 Mar 17;25(3):263-5. doi: 10.1016/j.ccr.2014.02.022. Cancer Cell. 2014. PMID: 24651006
-
Harnessing the PI3K/Akt/mTOR pathway in T-cell acute lymphoblastic leukemia: eliminating activity by targeting at different levels.Oncotarget. 2012 Aug;3(8):811-23. doi: 10.18632/oncotarget.579. Oncotarget. 2012. PMID: 22885370 Free PMC article.
-
Fishing for insight into leukemia relapse.Lab Anim (NY). 2014 Apr 21;43(5):151. doi: 10.1038/laban.534. Lab Anim (NY). 2014. PMID: 24751838 No abstract available.
-
NOTCH and phosphatidylinositide 3-kinase/phosphatase and tensin homolog deleted on chromosome ten/AKT/mammalian target of rapamycin (mTOR) signaling in T-cell development and T-cell acute lymphoblastic leukemia.Leuk Lymphoma. 2011 Jul;52(7):1200-10. doi: 10.3109/10428194.2011.564696. Epub 2011 Apr 4. Leuk Lymphoma. 2011. PMID: 21463127 Review.
-
Targeting signaling pathways in T-cell acute lymphoblastic leukemia initiating cells.Adv Biol Regul. 2014 Sep;56:6-21. doi: 10.1016/j.jbior.2014.04.004. Epub 2014 Apr 30. Adv Biol Regul. 2014. PMID: 24819383 Review.
Cited by
-
Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials.Nat Rev Drug Discov. 2021 Aug;20(8):611-628. doi: 10.1038/s41573-021-00210-8. Epub 2021 Jun 11. Nat Rev Drug Discov. 2021. PMID: 34117457 Free PMC article. Review.
-
Rictor has a pivotal role in maintaining quiescence as well as stemness of leukemia stem cells in MLL-driven leukemia.Leukemia. 2017 Feb;31(2):414-422. doi: 10.1038/leu.2016.223. Epub 2016 Aug 8. Leukemia. 2017. PMID: 27499138
-
Tackling Acute Lymphoblastic Leukemia-One Fish at a Time.Int J Mol Sci. 2019 Oct 25;20(21):5313. doi: 10.3390/ijms20215313. Int J Mol Sci. 2019. PMID: 31731471 Free PMC article. Review.
-
Modeling hematopoietic disorders in zebrafish.Dis Model Mech. 2019 Sep 6;12(9):dmm040360. doi: 10.1242/dmm.040360. Dis Model Mech. 2019. PMID: 31519693 Free PMC article. Review.
-
Therapeutic targeting of LCK tyrosine kinase and mTOR signaling in T-cell acute lymphoblastic leukemia.Blood. 2022 Oct 27;140(17):1891-1906. doi: 10.1182/blood.2021015106. Blood. 2022. PMID: 35544598 Free PMC article.
References
-
- Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277–302. - PubMed
-
- Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, Colman SM, Kempski H, Moorman AV, Titley I, Swansbury J, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. - PubMed
-
- Aparicio S, Caldas C. The Implications of Clonal Genome Evolution for Cancer Medicine. New England Journal of Medicine. 2013;368:842–851. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

