Evolution of the retroviral restriction gene Fv1: inhibition of non-MLV retroviruses
- PMID: 24603659
- PMCID: PMC3948346
- DOI: 10.1371/journal.ppat.1003968
Evolution of the retroviral restriction gene Fv1: inhibition of non-MLV retroviruses
Abstract
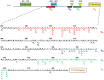
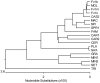
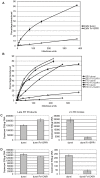
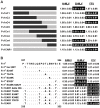
Fv1 is the prototypic restriction factor that protects against infection by the murine leukemia virus (MLV). It was first identified in cells that were derived from laboratory mice and was found to be homologous to the gag gene of an endogenous retrovirus (ERV). To understand the evolution of the host restriction gene from its retroviral origins, Fv1s from wild mice were isolated and characterized. Most of these possess intact open reading frames but not all restricted N-, B-, NR-or NB-tropic MLVs, suggesting that other viruses could have played a role in the selection of the gene. The Fv1s from Mus spretus and Mus caroli were found to restrict equine infectious anemia virus (EIAV) and feline foamy virus (FFV) respectively, indicating that Fv1 could have a broader target range than previously thought, including activity against lentiviruses and spumaviruses. Analyses of the Fv1 sequences revealed a number of residues in the C-terminal region that had evolved under positive selection. Four of these selected residues were found to be involved in the novel restriction by mapping studies. These results strengthen the similarities between the two capsid binding restriction factors, Fv1 and TRIM5α, which support the hypothesis that Fv1 defended mice against waves of retroviral infection possibly including non-MLVs as well as MLVs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Expression levels of Fv1: effects on retroviral restriction specificities.Retrovirology. 2016 Jun 24;13(1):42. doi: 10.1186/s12977-016-0276-7. Retrovirology. 2016. PMID: 27342974 Free PMC article.
-
Duplication and divergence of the retrovirus restriction gene Fv1 in Mus caroli allows protection from multiple retroviruses.PLoS Genet. 2020 Jun 11;16(6):e1008471. doi: 10.1371/journal.pgen.1008471. eCollection 2020 Jun. PLoS Genet. 2020. PMID: 32525879 Free PMC article.
-
Complex determinants within the Moloney murine leukemia virus capsid modulate susceptibility of the virus to Fv1 and Ref1-mediated restriction.Virology. 2007 Jul 5;363(2):245-55. doi: 10.1016/j.virol.2006.09.048. Epub 2007 Mar 6. Virology. 2007. PMID: 17343889
-
Post-entry restriction of retroviral infections.AIDS Rev. 2003 Jul-Sep;5(3):156-64. AIDS Rev. 2003. PMID: 14598564 Review.
-
Restriction factors: a defense against retroviral infection.Trends Microbiol. 2003 Jun;11(6):286-91. doi: 10.1016/s0966-842x(03)00123-9. Trends Microbiol. 2003. PMID: 12823946 Review.
Cited by
-
Transposable Element Domestication As an Adaptation to Evolutionary Conflicts.Trends Genet. 2017 Nov;33(11):817-831. doi: 10.1016/j.tig.2017.07.011. Epub 2017 Aug 24. Trends Genet. 2017. PMID: 28844698 Free PMC article. Review.
-
Foamy Viruses, Bet, and APOBEC3 Restriction.Viruses. 2021 Mar 18;13(3):504. doi: 10.3390/v13030504. Viruses. 2021. PMID: 33803830 Free PMC article. Review.
-
Mammalian genome innovation through transposon domestication.Nat Cell Biol. 2022 Sep;24(9):1332-1340. doi: 10.1038/s41556-022-00970-4. Epub 2022 Aug 25. Nat Cell Biol. 2022. PMID: 36008480 Free PMC article. Review.
-
Reconstruction of a replication-competent ancestral murine endogenous retrovirus-L.Retrovirology. 2018 May 2;15(1):34. doi: 10.1186/s12977-018-0416-3. Retrovirology. 2018. PMID: 29716624 Free PMC article.
-
Patterns of Coevolutionary Adaptations across Time and Space in Mouse Gammaretroviruses and Three Restrictive Host Factors.Viruses. 2021 Sep 18;13(9):1864. doi: 10.3390/v13091864. Viruses. 2021. PMID: 34578445 Free PMC article.
References
-
- Goff SP (2007) Retroviridae: The retroviruses and their replication. In: Knipe DM, Griffin DE, Lamb RA, Strauss SE, Howley, Marting MA, Roizman B, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins. Chapter 55: : pp1999–2069.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

