MLV requires Tap/NXF1-dependent pathway to export its unspliced RNA to the cytoplasm and to express both spliced and unspliced RNAs
- PMID: 24597485
- PMCID: PMC4015919
- DOI: 10.1186/1742-4690-11-21
MLV requires Tap/NXF1-dependent pathway to export its unspliced RNA to the cytoplasm and to express both spliced and unspliced RNAs
Abstract
Background: Eukaryotic cells have evolved stringent proofreading mechanisms to ensure that intron-containing mRNAs do not leave the nucleus. However, all retroviruses must bypass this checkpoint for replication. Indeed, their primary polycistronic transcript (Full-Length) must reach the cytoplasm to be either translated or packaged as genomic RNA in progeny viruses.Murine leukemia virus (MLV) is a prototype of simple retroviruses with only two well-regulated splicing events that directly influence viral leukemogenic properties in mice. Several cis-elements have been identified in the FL RNA that regulate its cytoplasmic accumulation. However, their connection with an export mechanism is yet unknown. Our goal was to identify the cellular pathway used by MLV to export its RNAs into the cytoplasm of the host cells.
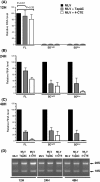
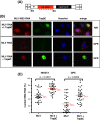
Results: Since other retroviruses use the CRM1 and/or the Tap/NXF1 pathways to export their unspliced RNA from the nucleus, we investigated the role of these two pathways in MLV replication by using specific inhibitors. The effects of export inhibition on MLV protein synthesis, RNA levels and RNA localization were studied by Western blotting, RT-qPCR, fluorescence microscopy and ribonucleoprotein immunoprecipitation assays. Taken together, our results show for the first time that MLV requires the Tap/NXF1-mediated export pathway, and not the CRM1 pathway, for the expression of its spliced and unspliced RNAs and for FL RNA nuclear export.
Conclusions: By contrast to HIV-1, MLV recruits the same pathway for the cytoplasmic expression of its spliced and unspliced RNAs. Thus, MLV RNA expression depends upon coordinated splicing/export processes. In addition, FL RNA translation relies on Tap/NXF1-dependent export, raising the critical question of whether the pool of FL RNA to be packaged is also exported by Tap/NXF1.
Figures


Similar articles
-
Strength in Diversity: Nuclear Export of Viral RNAs.Viruses. 2020 Sep 11;12(9):1014. doi: 10.3390/v12091014. Viruses. 2020. PMID: 32932882 Free PMC article. Review.
-
Murine leukemia virus uses NXF1 for nuclear export of spliced and unspliced viral transcripts.J Virol. 2014 Apr;88(8):4069-82. doi: 10.1128/JVI.03584-13. Epub 2014 Jan 29. J Virol. 2014. PMID: 24478440 Free PMC article.
-
NXF1 and CRM1 nuclear export pathways orchestrate nuclear export, translation and packaging of murine leukaemia retrovirus unspliced RNA.RNA Biol. 2020 Apr;17(4):528-538. doi: 10.1080/15476286.2020.1713539. Epub 2020 Jan 23. RNA Biol. 2020. PMID: 31918596 Free PMC article.
-
Insights into the nuclear export of murine leukemia virus intron-containing RNA.RNA Biol. 2015;12(9):942-9. doi: 10.1080/15476286.2015.1065375. RNA Biol. 2015. PMID: 26158194 Free PMC article. Review.
-
Murine leukemia virus uses TREX components for efficient nuclear export of unspliced viral transcripts.Viruses. 2014 Mar 10;6(3):1135-48. doi: 10.3390/v6031135. Viruses. 2014. PMID: 24618812 Free PMC article.
Cited by
-
Sequence and Functional Variation in the HIV-1 Rev Regulatory Axis.Curr HIV Res. 2020;18(2):85-98. doi: 10.2174/1570162X18666200106112842. Curr HIV Res. 2020. PMID: 31906839 Free PMC article.
-
From Cells to Virus Particles: Quantitative Methods to Monitor RNA Packaging.Viruses. 2016 Aug 22;8(8):239. doi: 10.3390/v8080239. Viruses. 2016. PMID: 27556480 Free PMC article. Review.
-
Diverse activities of viral cis-acting RNA regulatory elements revealed using multicolor, long-term, single-cell imaging.Mol Biol Cell. 2017 Feb 1;28(3):476-487. doi: 10.1091/mbc.E16-08-0612. Epub 2016 Nov 30. Mol Biol Cell. 2017. PMID: 27903772 Free PMC article.
-
Strength in Diversity: Nuclear Export of Viral RNAs.Viruses. 2020 Sep 11;12(9):1014. doi: 10.3390/v12091014. Viruses. 2020. PMID: 32932882 Free PMC article. Review.
-
Intron retention in viruses and cellular genes: Detention, border controls and passports.Wiley Interdiscip Rev RNA. 2018 May;9(3):e1470. doi: 10.1002/wrna.1470. Epub 2018 Mar 6. Wiley Interdiscip Rev RNA. 2018. PMID: 29508942 Free PMC article. Review.
References
-
- Bray M, Prasad S, Dubay JW, Hunter E, Jeang KT, Rekosh D, Hammarskjold ML. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev- independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

