Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology
- PMID: 24591590
- PMCID: PMC3964051
- DOI: 10.1073/pnas.1320670111
Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology
Abstract
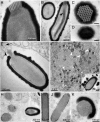
The largest known DNA viruses infect Acanthamoeba and belong to two markedly different families. The Megaviridae exhibit pseudo-icosahedral virions up to 0.7 μm in diameter and adenine-thymine (AT)-rich genomes of up to 1.25 Mb encoding a thousand proteins. Like their Mimivirus prototype discovered 10 y ago, they entirely replicate within cytoplasmic virion factories. In contrast, the recently discovered Pandoraviruses exhibit larger amphora-shaped virions 1 μm in length and guanine-cytosine-rich genomes up to 2.8 Mb long encoding up to 2,500 proteins. Their replication involves the host nucleus. Whereas the Megaviridae share some general features with the previously described icosahedral large DNA viruses, the Pandoraviruses appear unrelated to them. Here we report the discovery of a third type of giant virus combining an even larger pandoravirus-like particle 1.5 μm in length with a surprisingly smaller 600 kb AT-rich genome, a gene content more similar to Iridoviruses and Marseillevirus, and a fully cytoplasmic replication reminiscent of the Megaviridae. This suggests that pandoravirus-like particles may be associated with a variety of virus families more diverse than previously envisioned. This giant virus, named Pithovirus sibericum, was isolated from a >30,000-y-old radiocarbon-dated sample when we initiated a survey of the virome of Siberian permafrost. The revival of such an ancestral amoeba-infecting virus used as a safe indicator of the possible presence of pathogenic DNA viruses, suggests that the thawing of permafrost either from global warming or industrial exploitation of circumpolar regions might not be exempt from future threats to human or animal health.
Keywords: giant DNA virus; icosahedral capsid; late Pleistocene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba.Proc Natl Acad Sci U S A. 2015 Sep 22;112(38):E5327-35. doi: 10.1073/pnas.1510795112. Epub 2015 Sep 8. Proc Natl Acad Sci U S A. 2015. PMID: 26351664 Free PMC article.
-
Characterization of Mollivirus kamchatka, the First Modern Representative of the Proposed Molliviridae Family of Giant Viruses.J Virol. 2020 Mar 31;94(8):e01997-19. doi: 10.1128/JVI.01997-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996429 Free PMC article.
-
A Puzzling Anomaly in the 4-Mer Composition of the Giant Pandoravirus Genomes Reveals a Stringent New Evolutionary Selection Process.J Virol. 2019 Nov 13;93(23):e01206-19. doi: 10.1128/JVI.01206-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31534042 Free PMC article.
-
Viruses in close associations with free-living amoebae.Parasitol Res. 2015 Nov;114(11):3959-67. doi: 10.1007/s00436-015-4731-5. Epub 2015 Sep 16. Parasitol Res. 2015. PMID: 26374538 Review.
-
Looking at protists as a source of pathogenic viruses.Microb Pathog. 2014 Dec;77:131-5. doi: 10.1016/j.micpath.2014.09.005. Epub 2014 Sep 12. Microb Pathog. 2014. PMID: 25218687 Review.
Cited by
-
Quantitative conversion of biomass in giant DNA virus infection.Sci Rep. 2021 Mar 3;11(1):5025. doi: 10.1038/s41598-021-83547-9. Sci Rep. 2021. PMID: 33658544 Free PMC article.
-
Genetic manipulation of giant viruses and their host, Acanthamoeba castellanii.Nat Protoc. 2024 Jan;19(1):3-29. doi: 10.1038/s41596-023-00910-y. Epub 2023 Nov 14. Nat Protoc. 2024. PMID: 37964008 Review.
-
Medusavirus Ancestor in a Proto-Eukaryotic Cell: Updating the Hypothesis for the Viral Origin of the Nucleus.Front Microbiol. 2020 Sep 3;11:571831. doi: 10.3389/fmicb.2020.571831. eCollection 2020. Front Microbiol. 2020. PMID: 33013805 Free PMC article.
-
Comparison of a Modern and Fossil Pithovirus Reveals Its Genetic Conservation and Evolution.Genome Biol Evol. 2016 Aug 25;8(8):2333-9. doi: 10.1093/gbe/evw153. Genome Biol Evol. 2016. PMID: 27389688 Free PMC article.
-
Frozen Zoo: a collection of permafrost samples containing viable protists and their viruses.Biodivers Data J. 2020 Jul 10;8:e51586. doi: 10.3897/BDJ.8.e51586. eCollection 2020. Biodivers Data J. 2020. PMID: 32733138 Free PMC article.
References
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

