CRISPR-Cas systems for editing, regulating and targeting genomes
- PMID: 24584096
- PMCID: PMC4022601
- DOI: 10.1038/nbt.2842
CRISPR-Cas systems for editing, regulating and targeting genomes
Abstract
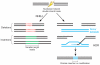
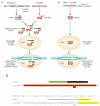
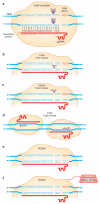
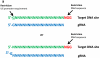
Targeted genome editing using engineered nucleases has rapidly gone from being a niche technology to a mainstream method used by many biological researchers. This widespread adoption has been largely fueled by the emergence of the clustered, regularly interspaced, short palindromic repeat (CRISPR) technology, an important new approach for generating RNA-guided nucleases, such as Cas9, with customizable specificities. Genome editing mediated by these nucleases has been used to rapidly, easily and efficiently modify endogenous genes in a wide variety of biomedically important cell types and in organisms that have traditionally been challenging to manipulate genetically. Furthermore, a modified version of the CRISPR-Cas9 system has been developed to recruit heterologous domains that can regulate endogenous gene expression or label specific genomic loci in living cells. Although the genome-wide specificities of CRISPR-Cas9 systems remain to be fully defined, the power of these systems to perform targeted, highly efficient alterations of genome sequence and gene expression will undoubtedly transform biological research and spur the development of novel molecular therapeutics for human disease.
Figures

Similar articles
-
Recent advances in CRISPR/Cas9 mediated genome editing in Bacillus subtilis.World J Microbiol Biotechnol. 2018 Sep 29;34(10):153. doi: 10.1007/s11274-018-2537-1. World J Microbiol Biotechnol. 2018. PMID: 30269229 Review.
-
Genome engineering via TALENs and CRISPR/Cas9 systems: challenges and perspectives.Plant Biotechnol J. 2014 Oct;12(8):1006-14. doi: 10.1111/pbi.12256. Plant Biotechnol J. 2014. PMID: 25250853 Review.
-
Selection and Validation of Spacer Sequences for CRISPR-Cas9 Genome Editing and Transcription Regulation in Bacteria.Methods Mol Biol. 2015;1334:233-44. doi: 10.1007/978-1-4939-2877-4_15. Methods Mol Biol. 2015. PMID: 26404154
-
Gene targeting technologies in rats: zinc finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats.Dev Growth Differ. 2014 Jan;56(1):46-52. doi: 10.1111/dgd.12110. Epub 2013 Dec 27. Dev Growth Differ. 2014. PMID: 24372523 Review.
-
How specific is CRISPR/Cas9 really?Curr Opin Chem Biol. 2015 Dec;29:72-8. doi: 10.1016/j.cbpa.2015.10.001. Epub 2015 Oct 24. Curr Opin Chem Biol. 2015. PMID: 26517564 Free PMC article. Review.
Cited by
-
Imaging of nanoparticle uptake and kinetics of intracellular trafficking in individual cells.Nanoscale. 2021 Jun 17;13(23):10436-10446. doi: 10.1039/d1nr00901j. Nanoscale. 2021. PMID: 34076024 Free PMC article.
-
Genetic Moderation of Stress Effects on Corticolimbic Circuitry.Neuropsychopharmacology. 2016 Jan;41(1):275-96. doi: 10.1038/npp.2015.216. Epub 2015 Jul 20. Neuropsychopharmacology. 2016. PMID: 26189450 Free PMC article. Review.
-
Genetic modification of the ant Lasius niger using CRISPR-Cas9 technology.Insect Mol Biol. 2023 Feb;32(1):11-25. doi: 10.1111/imb.12809. Epub 2022 Sep 9. Insect Mol Biol. 2023. PMID: 36030521 Free PMC article.
-
A versatile reporter system for CRISPR-mediated chromosomal rearrangements.Genome Biol. 2015 May 28;16(1):111. doi: 10.1186/s13059-015-0680-7. Genome Biol. 2015. PMID: 26018130 Free PMC article.
-
Postnatal therapeutic approaches in genetic neurodevelopmental disorders.Neural Regen Res. 2021 Mar;16(3):414-422. doi: 10.4103/1673-5374.293133. Neural Regen Res. 2021. PMID: 32985459 Free PMC article. Review.
References
-
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. - PubMed
-
- Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–209. - PubMed
-
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. - PubMed
-
- Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

