Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer
- PMID: 24581491
- PMCID: PMC3982296
- DOI: 10.1016/j.cell.2013.12.043
Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer
Abstract
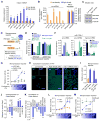
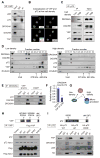
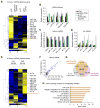
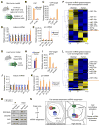
Global downregulation of microRNAs (miRNAs) is commonly observed in human cancers and can have a causative role in tumorigenesis. The mechanisms responsible for this phenomenon remain poorly understood. Here, we show that YAP, the downstream target of the tumor-suppressive Hippo-signaling pathway regulates miRNA biogenesis in a cell-density-dependent manner. At low cell density, nuclear YAP binds and sequesters p72 (DDX17), a regulatory component of the miRNA-processing machinery. At high cell density, Hippo-mediated cytoplasmic retention of YAP facilitates p72 association with Microprocessor and binding to a specific sequence motif in pri-miRNAs. Inactivation of the Hippo pathway or expression of constitutively active YAP causes widespread miRNA suppression in cells and tumors and a corresponding posttranscriptional induction of MYC expression. Thus, the Hippo pathway links contact-inhibition regulation to miRNA biogenesis and may be responsible for the widespread miRNA repression observed in cancer.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Tumour suppressors: Hippo promotes microRNA processing.Nat Rev Cancer. 2014 Apr;14(4):216-7. doi: 10.1038/nrc3715. Nat Rev Cancer. 2014. PMID: 24658271 No abstract available.
Similar articles
-
The Hippo pathway effectors TAZ/YAP regulate dicer expression and microRNA biogenesis through Let-7.J Biol Chem. 2014 Jan 24;289(4):1886-91. doi: 10.1074/jbc.C113.529362. Epub 2013 Dec 9. J Biol Chem. 2014. PMID: 24324261 Free PMC article.
-
Mutant p53 inhibits miRNA biogenesis by interfering with the microprocessor complex.Oncogene. 2016 Jul 21;35(29):3760-70. doi: 10.1038/onc.2016.51. Epub 2016 Mar 21. Oncogene. 2016. PMID: 26996669
-
Transcription and processing: multilayer controls of RNA biogenesis by the Hippo pathway.EMBO J. 2014 May 2;33(9):942-4. doi: 10.1002/embj.201488329. Epub 2014 Mar 17. EMBO J. 2014. PMID: 24639555 Free PMC article.
-
Crosstalk between Hippo signalling and miRNAs in tumour progression.FEBS J. 2017 Apr;284(7):1045-1055. doi: 10.1111/febs.13985. Epub 2017 Jan 21. FEBS J. 2017. PMID: 27973704 Review.
-
miRNAs and the Hippo pathway in cancer: Exploring the therapeutic potential (Review).Oncol Rep. 2022 Jul;48(1):135. doi: 10.3892/or.2022.8346. Epub 2022 Jun 14. Oncol Rep. 2022. PMID: 35699111 Review.
Cited by
-
New Insights into YES-Associated Protein Signaling Pathways in Hematological Malignancies: Diagnostic and Therapeutic Challenges.Cancers (Basel). 2021 Apr 20;13(8):1981. doi: 10.3390/cancers13081981. Cancers (Basel). 2021. PMID: 33924049 Free PMC article. Review.
-
Evidence for Functional Roles of MicroRNAs in Lineage Specification During Mouse and Human Preimplantation Development.Yale J Biol Med. 2023 Dec 29;96(4):481-494. doi: 10.59249/FOSI4358. eCollection 2023 Dec. Yale J Biol Med. 2023. PMID: 38161584 Free PMC article. Review.
-
MicroRNA biogenesis pathways in cancer.Nat Rev Cancer. 2015 Jun;15(6):321-33. doi: 10.1038/nrc3932. Nat Rev Cancer. 2015. PMID: 25998712 Free PMC article. Review.
-
Contact inhibition controls cell survival and proliferation via YAP/TAZ-autophagy axis.Nat Commun. 2018 Jul 27;9(1):2961. doi: 10.1038/s41467-018-05388-x. Nat Commun. 2018. PMID: 30054475 Free PMC article.
-
Implication of genetic variants in primary microRNA processing sites in the risk of multiple sclerosis.EBioMedicine. 2022 Jun;80:104052. doi: 10.1016/j.ebiom.2022.104052. Epub 2022 May 10. EBioMedicine. 2022. PMID: 35561450 Free PMC article.
References
-
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

