Dynamic survey of mitochondria by ubiquitin
- PMID: 24569520
- PMCID: PMC3989689
- DOI: 10.1002/embr.201338225
Dynamic survey of mitochondria by ubiquitin
Abstract
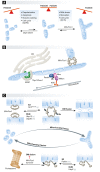
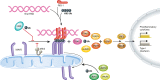
Ubiquitin is a post-translational modifier with proteolytic and non-proteolytic roles in many biological processes. At mitochondria, it performs regulatory homeostatic functions and contributes to mitochondrial quality control. Ubiquitin is essential for mitochondrial fusion, regulates mitochondria-ER contacts, and participates in maternal mtDNA inheritance. Under stress, mitochondrial dysfunction induces ubiquitin-dependent responses that involve mitochondrial proteome remodeling and culminate in organelle removal by mitophagy. In addition, many ubiquitin-dependent mechanisms have been shown to regulate innate immune responses and xenophagy. Here, we review the emerging roles of ubiquitin at mitochondria.
Figures

Similar articles
-
Regulation of mitochondrial cargo-selective autophagy by posttranslational modifications.J Biol Chem. 2021 Nov;297(5):101339. doi: 10.1016/j.jbc.2021.101339. Epub 2021 Oct 22. J Biol Chem. 2021. PMID: 34688664 Free PMC article. Review.
-
Quality control of the mitochondrion.Dev Cell. 2021 Apr 5;56(7):881-905. doi: 10.1016/j.devcel.2021.02.009. Epub 2021 Mar 3. Dev Cell. 2021. PMID: 33662258 Review.
-
Ubiquitin-mediated mitochondrial regulation by MITOL/MARCHF5 at a glance.J Biochem. 2022 Dec 27;173(1):1-11. doi: 10.1093/jb/mvac092. J Biochem. 2022. PMID: 36346121
-
Deubiquitinating enzymes regulate PARK2-mediated mitophagy.Autophagy. 2015 Apr 3;11(4):595-606. doi: 10.1080/15548627.2015.1034408. Autophagy. 2015. PMID: 25915564 Free PMC article.
-
Fndc-1 contributes to paternal mitochondria elimination in C. elegans.Dev Biol. 2019 Oct 1;454(1):15-20. doi: 10.1016/j.ydbio.2019.06.016. Epub 2019 Jun 21. Dev Biol. 2019. PMID: 31233739 Free PMC article.
Cited by
-
Role of Mitofusins and Mitophagy in Life or Death Decisions.Front Cell Dev Biol. 2020 Sep 22;8:572182. doi: 10.3389/fcell.2020.572182. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33072754 Free PMC article. Review.
-
Clearing tail-anchored proteins from mitochondria.Proc Natl Acad Sci U S A. 2014 Jun 3;111(22):7888-9. doi: 10.1073/pnas.1406864111. Epub 2014 May 22. Proc Natl Acad Sci U S A. 2014. PMID: 24853504 Free PMC article. No abstract available.
-
Pharmacological Progress of Mitophagy Regulation.Curr Neuropharmacol. 2023;21(5):1026-1041. doi: 10.2174/1570159X21666230314140528. Curr Neuropharmacol. 2023. PMID: 36918785 Free PMC article.
-
Mitochondrial Quality Control in Age-Related Pulmonary Fibrosis.Int J Mol Sci. 2020 Jan 18;21(2):643. doi: 10.3390/ijms21020643. Int J Mol Sci. 2020. PMID: 31963720 Free PMC article. Review.
-
Regulators of mitochondrial dynamics in cancer.Curr Opin Cell Biol. 2016 Apr;39:43-52. doi: 10.1016/j.ceb.2016.02.001. Epub 2016 Feb 18. Curr Opin Cell Biol. 2016. PMID: 26896558 Free PMC article. Review.
References
-
- Schwartz DC, Hochstrasser M. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem Sci. 2003;28:321–328. - PubMed
-
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. - PubMed
-
- Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. - PubMed
-
- Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

