FISH Amyloid - a new method for finding amyloidogenic segments in proteins based on site specific co-occurrence of aminoacids
- PMID: 24564523
- PMCID: PMC3941796
- DOI: 10.1186/1471-2105-15-54
FISH Amyloid - a new method for finding amyloidogenic segments in proteins based on site specific co-occurrence of aminoacids
Abstract
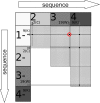
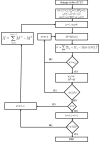
Background: Amyloids are proteins capable of forming fibrils whose intramolecular contact sites assume densely packed zipper pattern. Their oligomers can underlie serious diseases, e.g. Alzheimer's and Parkinson's diseases. Recent studies show that short segments of aminoacids can be responsible for amyloidogenic properties of a protein. A few hundreds of such peptides have been experimentally found but experimental testing of all candidates is currently not feasible. Here we propose an original machine learning method for classification of aminoacid sequences, based on discovering a segment with a discriminative pattern of site-specific co-occurrences between sequence elements. The pattern is based on the positions of residues with correlated occurrence over a sliding window of a specified length. The algorithm first recognizes the most relevant training segment in each positive training instance. Then the classification is based on maximal distances between co-occurrence matrix of the relevant segments in positive training sequences and the matrix from negative training segments. The method was applied for studying sequences of aminoacids with regard to their amyloidogenic properties.
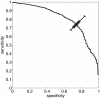
Results: Our method was first trained on available datasets of hexapeptides with the amyloidogenic classification, using 5 or 6-residue sliding windows. Depending on the choice of training and testing datasets, the area under ROC curve obtained the value up to 0.80 for experimental, and 0.95 for computationally generated (with 3D profile method) datasets. Importantly, the results on 5-residue segments were not significantly worse, although the classification required that algorithm first recognized the most relevant training segments. The dataset of long sequences, such as sup35 prion and a few other amyloid proteins, were applied to test the method and gave encouraging results. Our web tool FISH Amyloid was trained on all available experimental data 4-10 residues long, offers prediction of amyloidogenic segments in protein sequences.
Conclusions: We proposed a new original classification method which recognizes co-occurrence patterns in sequences. The method reveals characteristic classification pattern of the data and finds the segments where its scoring is the strongest, also in long training sequences. Applied to the problem of amyloidogenic segments recognition, it showed a good potential for classification problems in bioinformatics.
Figures


Similar articles
-
Machine learning methods can replace 3D profile method in classification of amyloidogenic hexapeptides.BMC Bioinformatics. 2013 Jan 17;14:21. doi: 10.1186/1471-2105-14-21. BMC Bioinformatics. 2013. PMID: 23327628 Free PMC article.
-
On the amyloid datasets used for training PAFIG--how (not) to extend the experimental dataset of hexapeptides.BMC Bioinformatics. 2013 Dec 4;14:351. doi: 10.1186/1471-2105-14-351. BMC Bioinformatics. 2013. PMID: 24305169 Free PMC article.
-
Exploiting heterogeneous features to improve in silico prediction of peptide status - amyloidogenic or non-amyloidogenic.BMC Bioinformatics. 2011;12 Suppl 13(Suppl 13):S21. doi: 10.1186/1471-2105-12-S13-S21. Epub 2011 Nov 30. BMC Bioinformatics. 2011. PMID: 22373069 Free PMC article.
-
Computational Approaches to Identification of Aggregation Sites and the Mechanism of Amyloid Growth.Adv Exp Med Biol. 2015;855:213-39. doi: 10.1007/978-3-319-17344-3_9. Adv Exp Med Biol. 2015. PMID: 26149932 Review.
-
Amino Acid Encoding Methods for Protein Sequences: A Comprehensive Review and Assessment.IEEE/ACM Trans Comput Biol Bioinform. 2020 Nov-Dec;17(6):1918-1931. doi: 10.1109/TCBB.2019.2911677. Epub 2020 Dec 8. IEEE/ACM Trans Comput Biol Bioinform. 2020. PMID: 30998480 Review.
Cited by
-
AggreProt: a web server for predicting and engineering aggregation prone regions in proteins.Nucleic Acids Res. 2024 Jul 5;52(W1):W159-W169. doi: 10.1093/nar/gkae420. Nucleic Acids Res. 2024. PMID: 38801076 Free PMC article.
-
Identification of fibrillogenic regions in human triosephosphate isomerase.PeerJ. 2016 Feb 4;4:e1676. doi: 10.7717/peerj.1676. eCollection 2016. PeerJ. 2016. PMID: 26870617 Free PMC article.
-
Assessment of Therapeutic Antibody Developability by Combinations of In Vitro and In Silico Methods.Methods Mol Biol. 2022;2313:57-113. doi: 10.1007/978-1-0716-1450-1_4. Methods Mol Biol. 2022. PMID: 34478132
-
Quantitating denaturation by formic acid: imperfect repeats are essential to the stability of the functional amyloid protein FapC.J Biol Chem. 2020 Sep 11;295(37):13031-13046. doi: 10.1074/jbc.RA120.013396. Epub 2020 Jul 21. J Biol Chem. 2020. PMID: 32719003 Free PMC article.
-
Amyloidogenic motifs revealed by n-gram analysis.Sci Rep. 2017 Oct 11;7(1):12961. doi: 10.1038/s41598-017-13210-9. Sci Rep. 2017. PMID: 29021608 Free PMC article.
References
-
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen AØ, Riekel C, Eisenberg D. Atomic structures of amyloid cross β-spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

