ISWI chromatin remodeling: one primary actor or a coordinated effort?
- PMID: 24561830
- PMCID: PMC4332870
- DOI: 10.1016/j.sbi.2014.01.010
ISWI chromatin remodeling: one primary actor or a coordinated effort?
Abstract
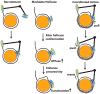
The ISWI family of ATP-dependent chromatin remodelers regulates transcription of coding and noncoding RNA by mobilizing nucleosomes and controlling the length of linker DNA separating nucleosomes (spacing). Nucleosome movement is tightly coupled to the DNA translocation activity of the helicase domain in the catalytic subunit. There may be other domains besides the helicase domain needed to move DNA in and out of nucleosomes. The C terminus of the ISWI catalytic subunit with the conserved HAND, SANT, and SLIDE domains may be involved in nucleosome spacing. There are several models of how the C terminus may facilitate in ISWI remodeling such as regulating the activity of the helicase domain and causing the helicase domain to translocate more efficiently on DNA or to enhance its selectivity for nucleosomes. Another possibility is that domains like SLIDE promote linker DNA entering into nucleosomes in a coordinated manner with the helicase domain.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
The ATPase domain of ISWI is an autonomous nucleosome remodeling machine.Nat Struct Mol Biol. 2013 Jan;20(1):82-9. doi: 10.1038/nsmb.2457. Epub 2012 Dec 2. Nat Struct Mol Biol. 2013. PMID: 23202585
-
Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes.Nature. 2012 Dec 13;492(7428):280-4. doi: 10.1038/nature11625. Epub 2012 Nov 11. Nature. 2012. PMID: 23143334 Free PMC article.
-
Low processivity for DNA translocation by the ISWI molecular motor.Biochim Biophys Acta. 2015 Oct;1854(10 Pt A):1487-93. doi: 10.1016/j.bbapap.2015.06.011. Epub 2015 Jun 25. Biochim Biophys Acta. 2015. PMID: 26116984
-
Mechanisms for nucleosome movement by ATP-dependent chromatin remodeling complexes.Results Probl Cell Differ. 2006;41:127-48. doi: 10.1007/400_005. Results Probl Cell Differ. 2006. PMID: 16909894 Review.
-
A proposal for kinetic proof reading by ISWI family chromatin remodeling motors.Curr Opin Chem Biol. 2010 Oct;14(5):660-5. doi: 10.1016/j.cbpa.2010.08.001. Epub 2010 Sep 15. Curr Opin Chem Biol. 2010. PMID: 20833099 Free PMC article. Review.
Cited by
-
Transcriptional co-activators: emerging roles in signaling pathways and potential therapeutic targets for diseases.Signal Transduct Target Ther. 2023 Nov 13;8(1):427. doi: 10.1038/s41392-023-01651-w. Signal Transduct Target Ther. 2023. PMID: 37953273 Free PMC article. Review.
-
Dual Recognition of H3K4me3 and DNA by the ISWI Component ARID5 Regulates the Floral Transition in Arabidopsis.Plant Cell. 2020 Jul;32(7):2178-2195. doi: 10.1105/tpc.19.00944. Epub 2020 Apr 30. Plant Cell. 2020. PMID: 32358072 Free PMC article.
-
The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families.Nat Commun. 2022 Dec 6;13(1):7524. doi: 10.1038/s41467-022-35002-0. Nat Commun. 2022. PMID: 36473839 Free PMC article.
-
On the role of inter-nucleosomal interactions and intrinsic nucleosome dynamics in chromatin function.Biochem Biophys Rep. 2016 Feb 16;5:492-501. doi: 10.1016/j.bbrep.2016.02.009. eCollection 2016 Mar. Biochem Biophys Rep. 2016. PMID: 28955857 Free PMC article. Review.
-
The ISWI remodeler in plants: protein complexes, biochemical functions, and developmental roles.Chromosoma. 2017 Jun;126(3):365-373. doi: 10.1007/s00412-017-0626-9. Epub 2017 Feb 17. Chromosoma. 2017. PMID: 28213686 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

