Radial symmetry in a chimeric glutamate receptor pore
- PMID: 24561802
- PMCID: PMC3962659
- DOI: 10.1038/ncomms4349
Radial symmetry in a chimeric glutamate receptor pore
Abstract
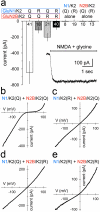
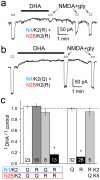
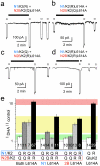
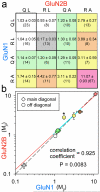
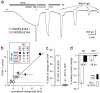
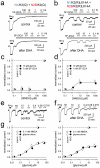
Ionotropic glutamate receptors comprise two conformationally different A/C and B/D subunit pairs. Closed channels exhibit fourfold radial symmetry in the transmembrane domain (TMD) but transition to twofold dimer-of-dimers symmetry for extracellular ligand binding and N-terminal domains. Here, to evaluate symmetry in open pores we analysed interaction between the Q/R editing site near the pore loop apex and the transmembrane M3 helix of kainate receptor subunit GluK2. Chimeric subunits that combined the GluK2 TMD with extracellular segments from NMDA receptors, which are obligate heteromers, yielded channels made up of A/C and B/D subunit pairs with distinct substitutions along M3 and/or Q/R site editing status, in an otherwise identical homotetrameric TMD. Our results indicate that Q/R site interaction with M3 occurs within individual subunits and is essentially the same for both A/C and B/D subunit conformations, suggesting that fourfold pore symmetry persists in the open state.
Figures

Similar articles
-
Chimeric Glutamate Receptor Subunits Reveal the Transmembrane Domain Is Sufficient for NMDA Receptor Pore Properties but Some Positive Allosteric Modulators Require Additional Domains.J Neurosci. 2016 Aug 24;36(34):8815-25. doi: 10.1523/JNEUROSCI.0345-16.2016. J Neurosci. 2016. PMID: 27559165 Free PMC article.
-
Fatty acid modulation and polyamine block of GluK2 kainate receptors analyzed by scanning mutagenesis.J Gen Physiol. 2010 Sep;136(3):339-52. doi: 10.1085/jgp.201010442. J Gen Physiol. 2010. PMID: 20805577 Free PMC article.
-
Neto proteins regulate gating of the kainate-type glutamate receptor GluK2 through two binding sites.J Biol Chem. 2019 Nov 22;294(47):17889-17902. doi: 10.1074/jbc.RA119.008631. Epub 2019 Oct 18. J Biol Chem. 2019. PMID: 31628192 Free PMC article.
-
Emerging structural insights into the function of ionotropic glutamate receptors.Trends Biochem Sci. 2015 Jun;40(6):328-37. doi: 10.1016/j.tibs.2015.04.002. Epub 2015 May 1. Trends Biochem Sci. 2015. PMID: 25941168 Free PMC article. Review.
-
Mammalian ionotropic glutamate receptors.Curr Opin Neurobiol. 1993 Jun;3(3):291-8. doi: 10.1016/0959-4388(93)90120-n. Curr Opin Neurobiol. 1993. PMID: 8396474 Review.
Cited by
-
Chimeric Glutamate Receptor Subunits Reveal the Transmembrane Domain Is Sufficient for NMDA Receptor Pore Properties but Some Positive Allosteric Modulators Require Additional Domains.J Neurosci. 2016 Aug 24;36(34):8815-25. doi: 10.1523/JNEUROSCI.0345-16.2016. J Neurosci. 2016. PMID: 27559165 Free PMC article.
-
Cadmium opens GluK2 kainate receptors with cysteine substitutions at the M3 helix bundle crossing.J Gen Physiol. 2019 Apr 1;151(4):435-451. doi: 10.1085/jgp.201812234. Epub 2018 Nov 29. J Gen Physiol. 2019. PMID: 30498132 Free PMC article.
-
Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels.Nat Struct Mol Biol. 2016 Jun 7;23(6):494-502. doi: 10.1038/nsmb.3214. Nat Struct Mol Biol. 2016. PMID: 27273633 Review.
-
Distinct GluN1 and GluN2 Structural Determinants for Subunit-Selective Positive Allosteric Modulation of N-Methyl-d-aspartate Receptors.ACS Chem Neurosci. 2021 Jan 6;12(1):79-98. doi: 10.1021/acschemneuro.0c00561. Epub 2020 Dec 16. ACS Chem Neurosci. 2021. PMID: 33326224 Free PMC article.
-
Two adjacent phenylalanines in the NMDA receptor GluN2A subunit M3 domain interactively regulate alcohol sensitivity and ion channel gating.Neuropharmacology. 2017 Mar 1;114:20-33. doi: 10.1016/j.neuropharm.2016.11.013. Epub 2016 Nov 19. Neuropharmacology. 2017. PMID: 27876530 Free PMC article.
References
-
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

