Evaluation of copy number variation detection for a SNP array platform
- PMID: 24555668
- PMCID: PMC4015297
- DOI: 10.1186/1471-2105-15-50
Evaluation of copy number variation detection for a SNP array platform
Abstract
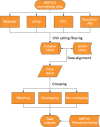
Background: Copy Number Variations (CNVs) are usually inferred from Single Nucleotide Polymorphism (SNP) arrays by use of some software packages based on given algorithms. However, there is no clear understanding of the performance of these software packages; it is therefore difficult to select one or several software packages for CNV detection based on the SNP array platform.We selected four publicly available software packages designed for CNV calling from an Affymetrix SNP array, including Birdsuite, dChip, Genotyping Console (GTC) and PennCNV. The publicly available dataset generated by Array-based Comparative Genomic Hybridization (CGH), with a resolution of 24 million probes per sample, was considered to be the "gold standard". Compared with the CGH-based dataset, the success rate, average stability rate, sensitivity, consistence and reproducibility of these four software packages were assessed compared with the "gold standard". Specially, we also compared the efficiency of detecting CNVs simultaneously by two, three and all of the software packages with that by a single software package.
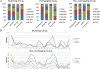
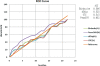
Results: Simply from the quantity of the detected CNVs, Birdsuite detected the most while GTC detected the least. We found that Birdsuite and dChip had obvious detecting bias. And GTC seemed to be inferior because of the least amount of CNVs it detected. Thereafter we investigated the detection consistency produced by one certain software package and the rest three software suits. We found that the consistency of dChip was the lowest while GTC was the highest. Compared with the CNVs detecting result of CGH, in the matching group, GTC called the most matching CNVs, PennCNV-Affy ranked second. In the non-overlapping group, GTC called the least CNVs. With regards to the reproducibility of CNV calling, larger CNVs were usually replicated better. PennCNV-Affy shows the best consistency while Birdsuite shows the poorest.
Conclusion: We found that PennCNV outperformed the other three packages in the sensitivity and specificity of CNV calling. Obviously, each calling method had its own limitations and advantages for different data analysis. Therefore, the optimized calling methods might be identified using multiple algorithms to evaluate the concordance and discordance of SNP array-based CNV calling.
Figures

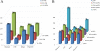

Similar articles
-
Accuracy of CNV Detection from GWAS Data.PLoS One. 2011 Jan 13;6(1):e14511. doi: 10.1371/journal.pone.0014511. PLoS One. 2011. PMID: 21249187 Free PMC article.
-
Software comparison for evaluating genomic copy number variation for Affymetrix 6.0 SNP array platform.BMC Bioinformatics. 2011 May 31;12:220. doi: 10.1186/1471-2105-12-220. BMC Bioinformatics. 2011. PMID: 21627824 Free PMC article.
-
Family-Based Benchmarking of Copy Number Variation Detection Software.PLoS One. 2015 Jul 21;10(7):e0133465. doi: 10.1371/journal.pone.0133465. eCollection 2015. PLoS One. 2015. PMID: 26197066 Free PMC article.
-
A survey of analysis software for array-comparative genomic hybridisation studies to detect copy number variation.Hum Genomics. 2010 Aug;4(6):421-7. doi: 10.1186/1479-7364-4-6-421. Hum Genomics. 2010. PMID: 20846932 Free PMC article. Review.
-
Copy number variations and stroke.Neurol Sci. 2016 Dec;37(12):1895-1904. doi: 10.1007/s10072-016-2658-y. Epub 2016 Jul 8. Neurol Sci. 2016. PMID: 27393281 Free PMC article. Review.
Cited by
-
Genome-wide detection of CNVs and their association with performance traits in broilers.BMC Genomics. 2021 May 17;22(1):354. doi: 10.1186/s12864-021-07676-1. BMC Genomics. 2021. PMID: 34001004 Free PMC article.
-
Data analysis in the post-genome-wide association study era.Chronic Dis Transl Med. 2016 Dec 21;2(4):231-234. doi: 10.1016/j.cdtm.2016.11.009. eCollection 2016 Dec. Chronic Dis Transl Med. 2016. PMID: 29063047 Free PMC article. Review.
-
A statistical approach to detection of copy number variations in PCR-enriched targeted sequencing data.BMC Bioinformatics. 2016 Oct 22;17(1):429. doi: 10.1186/s12859-016-1272-6. BMC Bioinformatics. 2016. PMID: 27770783 Free PMC article.
-
Bioinformatics Analysis for Circulating Cell-Free DNA in Cancer.Cancers (Basel). 2019 Jun 11;11(6):805. doi: 10.3390/cancers11060805. Cancers (Basel). 2019. PMID: 31212602 Free PMC article. Review.
-
Fully exploiting SNP arrays: a systematic review on the tools to extract underlying genomic structure.Brief Bioinform. 2022 Mar 10;23(2):bbac043. doi: 10.1093/bib/bbac043. Brief Bioinform. 2022. PMID: 35211719 Free PMC article.
References
-
- Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, Cole K, Mosse YP, Wood A, Lynch JE, Pecor K, Diamond M, Winter C, Wang K, Kim C, Geiger EA, McGrady PW, Blakemore AI, London WB, Shaikh TH, Bradfield J, Grant SF, Li H, Devoto M, Rappaport ER, Hakonarson H, Maris JM. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459(7249):987–991. doi: 10.1038/nature08035. - DOI - PMC - PubMed
-
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimäki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC. et al.Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. - DOI - PMC - PubMed
-
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, Murthy KK, Rovin BH, Bradley W, Clark RA, Anderson SA, O'connell RJ, Agan BK, Ahuja SS, Bologna R, Sen L, Dolan MJ, Ahuja SK. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307(5714):1434–1440. doi: 10.1126/science.1101160. - DOI - PubMed
-
- Kim J, Yim S, Jeong Y, Jung S, Xu H, Shin S, Chung Y. Comparison of normalization methods for defining copy number variation using whole-genome SNP genotyping data. Genomics Inf. 2008;6(4):231–234. doi: 10.5808/GI.2008.6.4.231. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

