Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila
- PMID: 24554766
- PMCID: PMC3982998
- DOI: 10.1091/mbc.E13-08-0449
Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila
Abstract
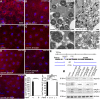
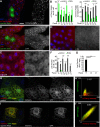
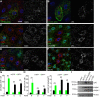
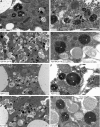
Homotypic fusion and vacuole protein sorting (HOPS) is a tethering complex required for trafficking to the vacuole/lysosome in yeast. Specific interaction of HOPS with certain SNARE (soluble NSF attachment protein receptor) proteins ensures the fusion of appropriate vesicles. HOPS function is less well characterized in metazoans. We show that all six HOPS subunits (Vps11 [vacuolar protein sorting 11]/CG32350, Vps18/Dor, Vps16A, Vps33A/Car, Vps39/CG7146, and Vps41/Lt) are required for fusion of autophagosomes with lysosomes in Drosophila. Loss of these genes results in large-scale accumulation of autophagosomes and blocks autophagic degradation under basal, starvation-induced, and developmental conditions. We find that HOPS colocalizes and interacts with Syntaxin 17 (Syx17), the recently identified autophagosomal SNARE required for fusion in Drosophila and mammals, suggesting their association is critical during tethering and fusion of autophagosomes with lysosomes. HOPS, but not Syx17, is also required for endocytic down-regulation of Notch and Boss in developing eyes and for proper trafficking to lysosomes and eye pigment granules. We also show that the formation of autophagosomes and their fusion with lysosomes is largely unaffected in null mutants of Vps38/UVRAG (UV radiation resistance associated), a suggested binding partner of HOPS in mammals, while endocytic breakdown and lysosome biogenesis is perturbed. Our results establish the role of HOPS and its likely mechanism of action during autophagy in metazoans.
Figures

Similar articles
-
The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17.Mol Biol Cell. 2014 Apr;25(8):1327-37. doi: 10.1091/mbc.E13-08-0447. Epub 2014 Feb 19. Mol Biol Cell. 2014. PMID: 24554770 Free PMC article.
-
Impairment of autophagosome-lysosome fusion in the buff mutant mice with the VPS33A(D251E) mutation.Autophagy. 2015;11(9):1608-22. doi: 10.1080/15548627.2015.1072669. Autophagy. 2015. PMID: 26259518 Free PMC article.
-
Loss of Drosophila Vps16A enhances autophagosome formation through reduced Tor activity.Autophagy. 2015;11(8):1209-15. doi: 10.1080/15548627.2015.1059559. Autophagy. 2015. PMID: 26061715 Free PMC article.
-
Evolutionarily conserved role and physiological relevance of a STX17/Syx17 (syntaxin 17)-containing SNARE complex in autophagosome fusion with endosomes and lysosomes.Autophagy. 2013 Oct;9(10):1642-6. doi: 10.4161/auto.25684. Epub 2013 Jul 22. Autophagy. 2013. PMID: 24113031 Review.
-
Syntaxin 16's Newly Deciphered Roles in Autophagy.Cells. 2019 Dec 17;8(12):1655. doi: 10.3390/cells8121655. Cells. 2019. PMID: 31861136 Free PMC article. Review.
Cited by
-
CUR5g, a novel autophagy inhibitor, exhibits potent synergistic anticancer effects with cisplatin against non-small-cell lung cancer.Cell Death Discov. 2022 Oct 31;8(1):435. doi: 10.1038/s41420-022-01217-9. Cell Death Discov. 2022. PMID: 36316321 Free PMC article.
-
The Interplay Between Autophagy and Regulated Necrosis.Antioxid Redox Signal. 2023 Mar;38(7-9):550-580. doi: 10.1089/ars.2022.0110. Epub 2022 Oct 12. Antioxid Redox Signal. 2023. PMID: 36053716 Free PMC article. Review.
-
Physiological functions of ULK1/2.J Mol Biol. 2024 Aug 1;436(15):168472. doi: 10.1016/j.jmb.2024.168472. Epub 2024 Feb 2. J Mol Biol. 2024. PMID: 38311233 Review.
-
The BEACH Domain Is Critical for Blue Cheese Function in a Spatial and Epistatic Autophagy Hierarchy.Front Cell Dev Biol. 2019 Aug 2;7:129. doi: 10.3389/fcell.2019.00129. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31428609 Free PMC article.
-
Arabidopsis HOPS subunit VPS41 carries out plant-specific roles in vacuolar transport and vegetative growth.Plant Physiol. 2022 Jun 27;189(3):1416-1434. doi: 10.1093/plphys/kiac167. Plant Physiol. 2022. PMID: 35417008 Free PMC article.
References
-
- Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

