Small-angle X-ray scattering-derived structure of the HIV-1 5' UTR reveals 3D tRNA mimicry
- PMID: 24550473
- PMCID: PMC3948283
- DOI: 10.1073/pnas.1319658111
Small-angle X-ray scattering-derived structure of the HIV-1 5' UTR reveals 3D tRNA mimicry
Abstract
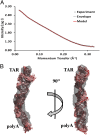
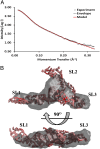
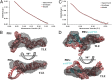
The most conserved region of the HIV type 1 (HIV-1) genome, the ∼335-nt 5' UTR, is characterized by functional stem loop domains responsible for regulating the viral life cycle. Despite the indispensable nature of this region of the genome in HIV-1 replication, 3D structures of multihairpin domains of the 5' UTR remain unknown. Using small-angle X-ray scattering and molecular dynamics simulations, we generated structural models of the transactivation (TAR)/polyadenylation (polyA), primer-binding site (PBS), and Psi-packaging domains. TAR and polyA form extended, coaxially stacked hairpins, consistent with their high stability and contribution to the pausing of reverse transcription. The Psi domain is extended, with each stem loop exposed for interactions with binding partners. The PBS domain adopts a bent conformation resembling the shape of a tRNA in apo and primer-annealed states. These results provide a structural basis for understanding several key molecular mechanisms underlying HIV-1 replication.
Keywords: HIV-1 RNA structure; molecular modeling; tRNA-like element.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Conservation of tRNA mimicry in the 5'-untranslated region of distinct HIV-1 subtypes.RNA. 2017 Dec;23(12):1850-1859. doi: 10.1261/rna.062182.117. Epub 2017 Aug 31. RNA. 2017. PMID: 28860303 Free PMC article.
-
A structured RNA motif is involved in correct placement of the tRNA(3)(Lys) primer onto the human immunodeficiency virus genome.J Virol. 2000 Mar;74(5):2227-38. doi: 10.1128/jvi.74.5.2227-2238.2000. J Virol. 2000. PMID: 10666253 Free PMC article.
-
Role of the 5' TAR stem--loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA.Biochemistry. 2008 Mar 11;47(10):3283-93. doi: 10.1021/bi7023173. Epub 2008 Feb 16. Biochemistry. 2008. PMID: 18278873
-
Structural determinants and mechanism of HIV-1 genome packaging.J Mol Biol. 2011 Jul 22;410(4):609-33. doi: 10.1016/j.jmb.2011.04.029. J Mol Biol. 2011. PMID: 21762803 Free PMC article. Review.
-
HIV-1 reverse transcription initiation: a potential target for novel antivirals?Virus Res. 2008 Jun;134(1-2):4-18. doi: 10.1016/j.virusres.2007.12.009. Epub 2008 Feb 6. Virus Res. 2008. PMID: 18255184 Review.
Cited by
-
Functional Equivalence of Retroviral MA Domains in Facilitating Psi RNA Binding Specificity by Gag.Viruses. 2016 Sep 19;8(9):256. doi: 10.3390/v8090256. Viruses. 2016. PMID: 27657107 Free PMC article.
-
Unpaired Guanosines in the 5' Untranslated Region of HIV-1 RNA Act Synergistically To Mediate Genome Packaging.J Virol. 2020 Oct 14;94(21):e00439-20. doi: 10.1128/JVI.00439-20. Print 2020 Oct 14. J Virol. 2020. PMID: 32796062 Free PMC article.
-
Pervasive tertiary structure in the dengue virus RNA genome.Proc Natl Acad Sci U S A. 2018 Nov 6;115(45):11513-11518. doi: 10.1073/pnas.1716689115. Epub 2018 Oct 19. Proc Natl Acad Sci U S A. 2018. PMID: 30341219 Free PMC article.
-
Rous Sarcoma Virus Genomic RNA Dimerization Capability In Vitro Is Not a Prerequisite for Viral Infectivity.Viruses. 2020 May 22;12(5):568. doi: 10.3390/v12050568. Viruses. 2020. PMID: 32455905 Free PMC article.
-
RNA structure determination: From 2D to 3D.Fundam Res. 2023 Jun 12;3(5):727-737. doi: 10.1016/j.fmre.2023.06.001. eCollection 2023 Sep. Fundam Res. 2023. PMID: 38933295 Free PMC article. Review.
References
-
- Bieniasz PD. An overview of intracellular interactions between immunodeficiency viruses and their hosts. AIDS. 2012;26(10):1243–1254. - PubMed
-
- Beerens N, Groot F, Berkhout B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J Biol Chem. 2001;276(33):31247–31256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

