Advantages of proteins being disordered
- PMID: 24532081
- PMCID: PMC4005706
- DOI: 10.1002/pro.2443
Advantages of proteins being disordered
Abstract
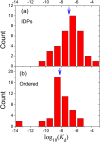
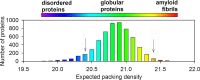
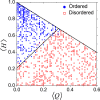
The past decade has witnessed great advances in our understanding of protein structure-function relationships in terms of the ubiquitous existence of intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs). The structural disorder of IDPs/IDRs enables them to play essential functions that are complementary to those of ordered proteins. In addition, IDPs/IDRs are persistent in evolution. Therefore, they are expected to possess some advantages over ordered proteins. In this review, we summarize and survey nine possible advantages of IDPs/IDRs: economizing genome/protein resources, overcoming steric restrictions in binding, achieving high specificity with low affinity, increasing binding rate, facilitating posttranslational modifications, enabling flexible linkers, preventing aggregation, providing resistance to non-native conditions, and allowing compatibility with more available sequences. Some potential advantages of IDPs/IDRs are not well understood and require both experimental and theoretical approaches to decipher. The connection with protein design is also briefly discussed.
Keywords: drug design; flexibility; intrinsically disordered proteins; molecular recognition; protein design; protein function; protein-protein interaction.
© 2014 The Protein Society.
Figures
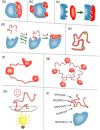
Similar articles
-
Features of molecular recognition of intrinsically disordered proteins via coupled folding and binding.Protein Sci. 2019 Nov;28(11):1952-1965. doi: 10.1002/pro.3718. Epub 2019 Sep 4. Protein Sci. 2019. PMID: 31441158 Free PMC article. Review.
-
Intrinsically disordered proteins/regions and insight into their biomolecular interactions.Biophys Chem. 2022 Apr;283:106769. doi: 10.1016/j.bpc.2022.106769. Epub 2022 Feb 1. Biophys Chem. 2022. PMID: 35139468 Review.
-
Conservation and coevolution determine evolvability of different classes of disordered residues in human intrinsically disordered proteins.Proteins. 2022 Mar;90(3):632-644. doi: 10.1002/prot.26261. Epub 2021 Oct 14. Proteins. 2022. PMID: 34626492
-
Functions of short lifetime biological structures at large: the case of intrinsically disordered proteins.Brief Funct Genomics. 2020 Jan 22;19(1):60-68. doi: 10.1093/bfgp/ely023. Brief Funct Genomics. 2020. PMID: 29982297 Review.
-
Do sequence neighbours of intrinsically disordered regions promote structural flexibility in intrinsically disordered proteins?J Struct Biol. 2020 Feb 1;209(2):107428. doi: 10.1016/j.jsb.2019.107428. Epub 2019 Nov 20. J Struct Biol. 2020. PMID: 31756456
Cited by
-
Structural dynamics shape the fitness window of alanine:glyoxylate aminotransferase.Protein Sci. 2022 May;31(5):e4303. doi: 10.1002/pro.4303. Protein Sci. 2022. PMID: 35481644 Free PMC article.
-
Portrait of the Intrinsically Disordered Side of the HTLV-1 Proteome.ACS Omega. 2019 Jun 7;4(6):10003-10018. doi: 10.1021/acsomega.9b01017. eCollection 2019 Jun 30. ACS Omega. 2019. PMID: 31460093 Free PMC article.
-
Advanced Sampling Methods for Multiscale Simulation of Disordered Proteins and Dynamic Interactions.Biomolecules. 2021 Sep 28;11(10):1416. doi: 10.3390/biom11101416. Biomolecules. 2021. PMID: 34680048 Free PMC article. Review.
-
Biophysics of protein evolution and evolutionary protein biophysics.J R Soc Interface. 2014 Nov 6;11(100):20140419. doi: 10.1098/rsif.2014.0419. J R Soc Interface. 2014. PMID: 25165599 Free PMC article.
-
Thermostable Proteins from HaCaT Keratinocytes Identify a Wide Breadth of Intrinsically Disordered Proteins and Candidates for Liquid-Liquid Phase Separation.Int J Mol Sci. 2022 Nov 18;23(22):14323. doi: 10.3390/ijms232214323. Int J Mol Sci. 2022. PMID: 36430801 Free PMC article.
References
-
- Tompa P. Intrinsically disordered proteins: a 10-year recap. Trends Biochem Sci. 2012;37:509–516. - PubMed
-
- Huang YQ, Liu ZR. Intrinsically disordered proteins: the new sequence-structure-function relations. Acta Phys Chim Sin. 2010;26:2061–2072.
-
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CR, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. - PubMed
-
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

