A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability
- PMID: 24528869
- PMCID: PMC3991421
- DOI: 10.1016/j.chom.2014.01.006
A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability
Abstract
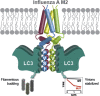
Autophagy recycles cellular components and defends cells against intracellular pathogens. While viruses must evade autophagocytic destruction, some viruses can also subvert autophagy for their own benefit. The ability of influenza A virus (IAV) to evade autophagy depends on the Matrix 2 (M2) ion-channel protein. We show that the cytoplasmic tail of IAV M2 interacts directly with the essential autophagy protein LC3 and promotes LC3 relocalization to the unexpected destination of the plasma membrane. LC3 binding is mediated by a highly conserved LC3-interacting region (LIR) in M2. The M2 LIR is required for LC3 redistribution to the plasma membrane in virus-infected cells. Mutations in M2 that abolish LC3 binding interfere with filamentous budding and reduce virion stability. IAV therefore subverts autophagy by mimicking a host short linear protein-protein interaction motif. This strategy may facilitate transmission of infection between organisms by enhancing the stability of viral progeny.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures
Comment in
-
Influenza A virus lures autophagic protein LC3 to budding sites.Cell Host Microbe. 2014 Feb 12;15(2):130-1. doi: 10.1016/j.chom.2014.01.014. Cell Host Microbe. 2014. PMID: 24528859
Similar articles
-
Influenza A virus lures autophagic protein LC3 to budding sites.Cell Host Microbe. 2014 Feb 12;15(2):130-1. doi: 10.1016/j.chom.2014.01.014. Cell Host Microbe. 2014. PMID: 24528859
-
Autophagy Promotes Replication of Influenza A Virus In Vitro.J Virol. 2019 Feb 5;93(4):e01984-18. doi: 10.1128/JVI.01984-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30541828 Free PMC article.
-
Influenza M2 protein regulates MAVS-mediated signaling pathway through interacting with MAVS and increasing ROS production.Autophagy. 2019 Jul;15(7):1163-1181. doi: 10.1080/15548627.2019.1580089. Epub 2019 Feb 20. Autophagy. 2019. PMID: 30741586 Free PMC article.
-
Influenza A Virus M2 Protein: Roles from Ingress to Egress.Int J Mol Sci. 2017 Dec 7;18(12):2649. doi: 10.3390/ijms18122649. Int J Mol Sci. 2017. PMID: 29215568 Free PMC article. Review.
-
Influenza virus assembly and budding.Virology. 2011 Mar 15;411(2):229-36. doi: 10.1016/j.virol.2010.12.003. Epub 2011 Jan 14. Virology. 2011. PMID: 21237476 Free PMC article. Review.
Cited by
-
The Emerging Roles of Viroporins in ER Stress Response and Autophagy Induction during Virus Infection.Viruses. 2015 Jun 4;7(6):2834-57. doi: 10.3390/v7062749. Viruses. 2015. PMID: 26053926 Free PMC article. Review.
-
White spot syndrome virus entry is dependent on multiple endocytic routes and strongly facilitated by Cq-GABARAP in a CME-dependent manner.Sci Rep. 2016 Jul 7;6:28694. doi: 10.1038/srep28694. Sci Rep. 2016. PMID: 27385304 Free PMC article.
-
Catch me if you can: the link between autophagy and viruses.PLoS Pathog. 2015 Mar 26;11(3):e1004685. doi: 10.1371/journal.ppat.1004685. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25811485 Free PMC article. Review. No abstract available.
-
Usp25-Erlin1/2 activity limits cholesterol flux to restrict virus infection.Dev Cell. 2023 Nov 20;58(22):2495-2509.e6. doi: 10.1016/j.devcel.2023.08.013. Epub 2023 Sep 7. Dev Cell. 2023. PMID: 37683630 Free PMC article.
-
Cathepsin B plays a key role in optimal production of the influenza A virus.J Virol Antivir Res. 2018 Apr;7(1):1-20. doi: 10.4172/2324-8955.1000178. Epub 2018 May 11. J Virol Antivir Res. 2018. PMID: 29349092 Free PMC article.
References
-
- Alemu E.A., Lamark T., Torgersen K.M., Birgisdottir A.B., Larsen K.B., Jain A., Olsvik H., Øvervatn A., Kirkin V., Johansen T. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 2012;287:39275–39290. - PMC - PubMed
-
- Barrios-Rodiles M., Brown K.R., Ozdamar B., Bose R., Liu Z., Donovan R.S., Shinjo F., Liu Y., Dembowy J., Taylor I.W. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. - PubMed
-
- Bourmakina S.V., García-Sastre A. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 2003;84:517–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

