Structure and function of human DnaJ homologue subfamily a member 1 (DNAJA1) and its relationship to pancreatic cancer
- PMID: 24512202
- PMCID: PMC3985919
- DOI: 10.1021/bi401329a
Structure and function of human DnaJ homologue subfamily a member 1 (DNAJA1) and its relationship to pancreatic cancer
Abstract
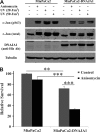
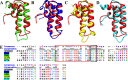
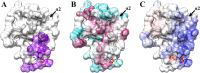
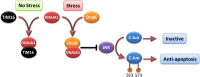
Pancreatic cancer has a dismal 5 year survival rate of 5.5% that has not been improved over the past 25 years despite an enormous amount of effort. Thus, there is an urgent need to identify truly novel yet druggable protein targets for drug discovery. The human protein DnaJ homologue subfamily A member 1 (DNAJA1) was previously shown to be downregulated 5-fold in pancreatic cancer cells and has been targeted as a biomarker for pancreatic cancer, but little is known about the specific biological function for DNAJA1 or the other members of the DnaJ family encoded in the human genome. Our results suggest the overexpression of DNAJA1 suppresses the stress response capabilities of the oncogenic transcription factor, c-Jun, and results in the diminution of cell survival. DNAJA1 likely activates a DnaK protein by forming a complex that suppresses the JNK pathway, the hyperphosphorylation of c-Jun, and the anti-apoptosis state found in pancreatic cancer cells. A high-quality nuclear magnetic resonance solution structure of the J-domain of DNAJA1 combined with a bioinformatics analysis and a ligand affinity screen identifies a potential DnaK binding site, which is also predicted to overlap with an inhibitory binding site, suggesting DNAJA1 activity is highly regulated.
Figures

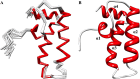


Similar articles
-
Leveraging the Structure of DNAJA1 to Discover Novel Potential Pancreatic Cancer Therapies.Biomolecules. 2022 Sep 29;12(10):1391. doi: 10.3390/biom12101391. Biomolecules. 2022. PMID: 36291603 Free PMC article.
-
Inhibition of mutant Kras and p53-driven pancreatic carcinogenesis by atorvastatin: Mainly via targeting of the farnesylated DNAJA1 in chaperoning mutant p53.Mol Carcinog. 2019 Nov;58(11):2052-2064. doi: 10.1002/mc.23097. Epub 2019 Aug 9. Mol Carcinog. 2019. PMID: 31397499 Free PMC article.
-
DNAJA1 Dysregulates Metabolism Promoting an Antiapoptotic Phenotype in Pancreatic Ductal Adenocarcinoma.J Proteome Res. 2021 Aug 6;20(8):3925-3939. doi: 10.1021/acs.jproteome.1c00233. Epub 2021 Jul 15. J Proteome Res. 2021. PMID: 34264680 Free PMC article.
-
Human DNAJ in cancer and stem cells.Cancer Lett. 2011 Dec 22;312(2):129-42. doi: 10.1016/j.canlet.2011.08.019. Epub 2011 Aug 27. Cancer Lett. 2011. PMID: 21925790 Review.
-
Multi-faceted role of HSP40 in cancer.Clin Exp Metastasis. 2009;26(6):559-67. doi: 10.1007/s10585-009-9255-x. Epub 2009 Apr 2. Clin Exp Metastasis. 2009. PMID: 19340594 Review.
Cited by
-
Dynamical Structures of Hsp70 and Hsp70-Hsp40 Complexes.Structure. 2016 Jul 6;24(7):1014-30. doi: 10.1016/j.str.2016.05.011. Epub 2016 Jun 23. Structure. 2016. PMID: 27345933 Free PMC article. Review.
-
Chemogenomic screening identifies the Hsp70 co-chaperone DNAJA1 as a hub for anticancer drug resistance.Sci Rep. 2020 Aug 14;10(1):13831. doi: 10.1038/s41598-020-70764-x. Sci Rep. 2020. PMID: 32796891 Free PMC article.
-
Environmental Stress Responses of DnaJA1, DnaJB12 and DnaJC8 in Apis cerana cerana.Front Genet. 2018 Oct 8;9:445. doi: 10.3389/fgene.2018.00445. eCollection 2018. Front Genet. 2018. PMID: 30349556 Free PMC article.
-
J domain independent functions of J proteins.Cell Stress Chaperones. 2016 Jul;21(4):563-70. doi: 10.1007/s12192-016-0697-1. Epub 2016 May 4. Cell Stress Chaperones. 2016. PMID: 27145962 Free PMC article. Review.
-
The Hsp70 co-chaperone Ydj1/HDJ2 regulates ribonucleotide reductase activity.PLoS Genet. 2018 Nov 19;14(11):e1007462. doi: 10.1371/journal.pgen.1007462. eCollection 2018 Nov. PLoS Genet. 2018. PMID: 30452489 Free PMC article.
References
-
- Hidalgo M. (2010) Pancreatic cancer. N. Engl. J. Med. 362, 1605–1617. - PubMed
-
- Siegel R.; Naishadham D.; Jemal A. (2012) Cancer statistics, 2012. Ca-Cancer J. Clin. 62, 10–29. - PubMed
-
- Li D.; Xie K.; Wolff R.; Abbruzzese J. L. (2004) Pancreatic cancer. Lancet 363, 1049–1057. - PubMed
-
- Hidalgo M. (2012) New insights into pancreatic cancer biology. Ann. Oncol. 23(Suppl. 10), x135–x138. - PubMed
-
- Mini E.; Nobili S.; Caciagli B.; Landini I.; Mazzei T. (2006) Cellular pharmacology of gemcitabine. Ann. Oncol. 17(Suppl. 5), v7–v12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

