High-throughput cloning and expression of integral membrane proteins in Escherichia coli
- PMID: 24510647
- PMCID: PMC3920300
- DOI: 10.1002/0471140864.ps2906s74
High-throughput cloning and expression of integral membrane proteins in Escherichia coli
Abstract
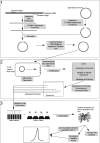
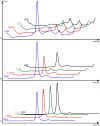
Recently, several structural genomics centers have been established and a remarkable number of three-dimensional structures of soluble proteins have been solved. For membrane proteins, the number of structures solved has been significantly trailing those for their soluble counterparts, not least because over-expression and purification of membrane proteins is a much more arduous process. By using high-throughput technologies, a large number of membrane protein targets can be screened simultaneously and a greater number of expression and purification conditions can be employed, leading to a higher probability of successfully determining the structure of membrane proteins. This unit describes the cloning, expression, and screening of membrane proteins using high-throughput methodologies developed in the laboratory. Basic Protocol 1 describes cloning of inserts into expression vectors by ligation-independent cloning. Basic Protocol 2 describes the expression and purification of the target proteins on a miniscale. Lastly, for the targets that do express on the miniscale, Basic Protocols 3 and 4 outline the methods employed for the expression and purification of targets on a midi-scale, as well as a procedure for detergent screening and identification of detergent(s) in which the target protein is stable.
Keywords: Escherichia coli; cloning/expression; high-throughput; ligation-independent cloning (LIC); membrane protein; membrane protein purification; recombinant protein expression.
Copyright © 2013 John Wiley & Sons, Inc.
Figures




Similar articles
-
An efficient strategy for high throughput screening of recombinant integral membrane protein expression and stability.Protein Expr Purif. 2011 Jul;78(1):6-13. doi: 10.1016/j.pep.2011.02.010. Epub 2011 Feb 24. Protein Expr Purif. 2011. PMID: 21354311
-
High-Throughput Cell-Free Screening of Eukaryotic Membrane Proteins in Lipidic Mimetics.Curr Protoc. 2022 Aug;2(8):e510. doi: 10.1002/cpz1.510. Curr Protoc. 2022. PMID: 35926131
-
High-throughput cloning, expression and purification of glycoside hydrolases using Ligation-Independent Cloning (LIC).Protein Expr Purif. 2014 Jul;99:35-42. doi: 10.1016/j.pep.2014.03.008. Epub 2014 Mar 26. Protein Expr Purif. 2014. PMID: 24680731
-
Expression, Solubilization, and Purification of Bacterial Membrane Proteins.Curr Protoc Protein Sci. 2016 Feb 2;83:29.15.1-29.15.15. doi: 10.1002/0471140864.ps2915s83. Curr Protoc Protein Sci. 2016. PMID: 26836409 Review.
-
Breaking the bottleneck: eukaryotic membrane protein expression for high-resolution structural studies.J Struct Biol. 2007 Dec;160(3):265-74. doi: 10.1016/j.jsb.2007.07.001. Epub 2007 Jul 14. J Struct Biol. 2007. PMID: 17702603 Review.
Cited by
-
High-throughput cell-free screening of eukaryotic membrane protein expression in lipidic mimetics.Protein Sci. 2022 Mar;31(3):639-651. doi: 10.1002/pro.4259. Epub 2021 Dec 23. Protein Sci. 2022. PMID: 34910339 Free PMC article.
-
Deuterium spin relaxation of fractionally deuterated ribonuclease H using paired 475 and 950 MHz NMR spectrometers.J Biomol NMR. 2024 Sep;78(3):169-177. doi: 10.1007/s10858-024-00443-w. Epub 2024 Jun 10. J Biomol NMR. 2024. PMID: 38856928
-
Atomic-level analysis of membrane-protein structure.Nat Struct Mol Biol. 2016 Jun 7;23(6):464-7. doi: 10.1038/nsmb.3215. Nat Struct Mol Biol. 2016. PMID: 27273628 Free PMC article. Review.
-
Fine Sampling of Sequence Space for Membrane Protein Structural Biology.J Mol Biol. 2021 Jul 23;433(15):167055. doi: 10.1016/j.jmb.2021.167055. Epub 2021 May 20. J Mol Biol. 2021. PMID: 34022208 Free PMC article.
-
Escherichia coli as host for membrane protein structure determination: a global analysis.Sci Rep. 2015 Jul 10;5:12097. doi: 10.1038/srep12097. Sci Rep. 2015. PMID: 26160693 Free PMC article.
References
Literature Cited
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Amann E, Brosius J. ATG vectors' for regulated high-level expression of cloned genes in Escherichia coli. Gene. 1985;40:183–190. - PubMed
-
- Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–1408. - PubMed
-
- Bernard HU, Helinski DR. Use of the lambda phage promoter PL to promote gene expression in hybrid plasmid cloning vehicles. Methods Enzymol. 1979;68:482–492. - PubMed
Key References
-
- Punta M, Love J, Handelman S, Hunt JF, Shapiro L, Hendrickson WA, Rost B. Structural genomics target selection for the New York consortium on membrane protein structure. J Struct Funct Genomics. 2009;10:255–268. Describes target selection of bacterial αIMPs for the NYCOMPS high through-put expression pipeline and discusses the potential impact of structure determination of these targets on a larger uncharacterized pool of proteins. - PMC - PubMed
-
- Kloppmann E, Punta M, Rost B. Structural genomics plucks high-hanging membrane proteins. Curr Opin Struct Biol. 2012;22:326–332. Reviews recent progress of IMP structure determination by structural genomics (SG) initiatives and discusses the progress of SG centers in covering important protein families. - PMC - PubMed
-
- Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming J, dhe-Paganon S, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau C, Kim R, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GS, Peleg Y, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SA, Wilson IA, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WH, Kim Y, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain K, Luz J, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan S, Studier FW, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, Hunt JF, Tong L, Cunningham K, Inouye M, Anderson S, Janjua H, Shastry R, Ho CK, Wang D, Wang H, Jiang M, Montelione GT, Stuart DI, Owens RJ, Daenke S, Schutz A, Heinemann U, Yokoyama S, Bussow K, Gunsalus KC. Protein production and purification. Nat Methods. 2008;5:135–146. - PMC - PubMed
-
- Stevens RC. Design of high-throughput methods of protein production for structural biology. Structure. 2000;8:R177–185. These two reviews describe the different cloning, expression and purification methods employed for high throughput protein production. The former also summarizes the different approaches used by structural genomics centers. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

