Proteomics analysis of cancer exosomes using a novel modified aptamer-based array (SOMAscan™) platform
- PMID: 24505114
- PMCID: PMC3977183
- DOI: 10.1074/mcp.M113.032136
Proteomics analysis of cancer exosomes using a novel modified aptamer-based array (SOMAscan™) platform
Abstract
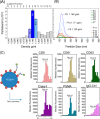
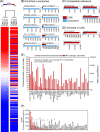
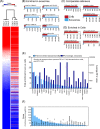
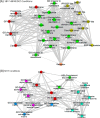
We have used a novel affinity-based proteomics technology to examine the protein signature of small secreted extracellular vesicles called exosomes. The technology uses a new class of protein binding reagents called SOMAmers® (slow off-rate modified aptamers) and allows the simultaneous precise measurement of over 1000 proteins. Exosomes were highly purified from the Du145 prostate cancer cell line, by pooling selected fractions from a continuous sucrose gradient (within the density range of 1.1 to 1.2 g/ml), and examined under standard conditions or with additional detergent treatment by the SOMAscan™ array (version 3.0). Lysates of Du145 cells were also prepared, and the profiles were compared. Housekeeping proteins such as cyclophilin-A, LDH, and Hsp70 were present in exosomes, and we identified almost 100 proteins that were enriched in exosomes relative to cells. These included proteins of known association with cancer exosomes such as MFG-E8, integrins, and MET, and also those less widely reported as exosomally associated, such as ROR1 and ITIH4. Several proteins with no previously known exosomal association were confirmed as exosomally expressed in experiments using individual SOMAmer® reagents or antibodies in micro-plate assays. Western blotting confirmed the SOMAscan™-identified enrichment of exosomal NOTCH-3, L1CAM, RAC1, and ADAM9. In conclusion, we describe here over 300 proteins of hitherto unknown association with prostate cancer exosomes and suggest that the SOMAmer®-based assay technology is an effective proteomics platform for exosome-associated biomarker discovery in diverse clinical settings.
Conflict of interest statement
Conflict of interest statement: Evaldas Katilius, Breanna C. Smith and Bridget Gordon are employed by SomaLogic Inc.
Figures

Similar articles
-
Proteomics analysis of bladder cancer exosomes.Mol Cell Proteomics. 2010 Jun;9(6):1324-38. doi: 10.1074/mcp.M000063-MCP201. Epub 2010 Mar 11. Mol Cell Proteomics. 2010. PMID: 20224111 Free PMC article.
-
Molecular profiling of prostate cancer derived exosomes may reveal a predictive signature for response to docetaxel.Oncotarget. 2015 Aug 28;6(25):21740-54. doi: 10.18632/oncotarget.3226. Oncotarget. 2015. PMID: 25844599 Free PMC article. Clinical Trial.
-
Proteomic profiling of exosomes leads to the identification of novel biomarkers for prostate cancer.PLoS One. 2013 Dec 31;8(12):e82589. doi: 10.1371/journal.pone.0082589. eCollection 2013. PLoS One. 2013. PMID: 24391718 Free PMC article.
-
Highly multiplexed proteomic platform for biomarker discovery, diagnostics, and therapeutics.Adv Exp Med Biol. 2013;735:283-300. doi: 10.1007/978-1-4614-4118-2_20. Adv Exp Med Biol. 2013. PMID: 23402035 Review.
-
Exosomal proteins as potential markers of tumor diagnosis.J Hematol Oncol. 2017 Dec 27;10(1):175. doi: 10.1186/s13045-017-0542-8. J Hematol Oncol. 2017. PMID: 29282096 Free PMC article. Review.
Cited by
-
Proteomics analysis of vesicles isolated from plasma and urine of prostate cancer patients using a multiplex, aptamer-based protein array.J Extracell Vesicles. 2016 Jun 29;5:31209. doi: 10.3402/jev.v5.31209. eCollection 2016. J Extracell Vesicles. 2016. PMID: 27363484 Free PMC article.
-
Identification of definitive serum biomarkers associated with disease activity in primary Sjögren's syndrome.Arthritis Res Ther. 2016 May 14;18(1):106. doi: 10.1186/s13075-016-1006-1. Arthritis Res Ther. 2016. PMID: 27180164 Free PMC article.
-
Plasma protein expression profiles, cardiovascular disease, and religious struggles among South Asians in the MASALA study.Sci Rep. 2021 Jan 13;11(1):961. doi: 10.1038/s41598-020-79429-1. Sci Rep. 2021. PMID: 33441605 Free PMC article.
-
Predicting the Uncertain Future of Aptamer-Based Diagnostics and Therapeutics.Molecules. 2015 Apr 16;20(4):6866-87. doi: 10.3390/molecules20046866. Molecules. 2015. PMID: 25913927 Free PMC article. Review.
-
Serum biomarkers associated with baseline clinical severity in young steroid-naïve Duchenne muscular dystrophy boys.Hum Mol Genet. 2020 Aug 29;29(15):2481-2495. doi: 10.1093/hmg/ddaa132. Hum Mol Genet. 2020. PMID: 32592467 Free PMC article. Clinical Trial.
References
-
- Thompson I. M., Pauler D. K., Goodman P. J., Tangen C. M., Lucia M. S., Parnes H. L., Minasian L. M., Ford L. G., Lippman S. M., Crawford E. D., Crowley J. J., Coltman C. A., Jr. (2004) Prevalence of prostate cancer among men with a prostate-specific antigen level ≤4.0 ng per milliliter. N. Engl. J. Med. 350, 2239–2246 - PubMed
-
- Selley S., Donovan J., Faulkner A., Coast J., Gillatt D. (1997) Diagnosis, management and screening of early localised prostate cancer. Health Technol. Assess. 1(2), 1–96 - PubMed
-
- Caby M., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. (2005) Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 17, 879–887 - PubMed
-
- Admyre C., Johansson S. M., Qazi K. R., Filen J.-J., Lahesmaa R., Norman M., Neve E. P., Scheynius A., Gabrielsson S. (2007) Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 179, 1969–1978 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

