DNA-PKcs is required to maintain stability of Chk1 and Claspin for optimal replication stress response
- PMID: 24500207
- PMCID: PMC3985680
- DOI: 10.1093/nar/gku116
DNA-PKcs is required to maintain stability of Chk1 and Claspin for optimal replication stress response
Abstract
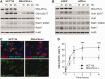
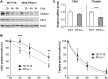
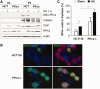
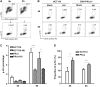
The ataxia telangiectasia mutated and Rad3-related (ATR)-checkpoint kinase 1 (Chk1) axis is the major signaling pathway activated in response to replication stress and is essential for the intra-S checkpoint. ATR phosphorylates and activates a number of molecules to coordinate cell cycle progression. Chk1 is the major effector downstream from ATR and plays a critical role in intra-S checkpoint on replication stress. Activation of Chk1 kinase also requires its association with Claspin, an adaptor protein essential for Chk1 protein stability, recruitment and ATR-dependent Chk1 phosphorylation. We have previously reported that, on replication stress, the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) is rapidly phosphorylated by ATR at the stalled replication forks and is required for cellular resistance to replication stresses although the impact of DNA-PKcs onto the ATR signaling pathway remains elusive. Here we report that ATR-dependent Chk1 phosphorylation and Chk1 signaling are compromised in the absence of DNA-PKcs. Our investigation reveals that DNA-PKcs is required to maintain Chk1-Claspin complex stability and transcriptional regulation of Claspin expression. The impaired Chk1 activity results in a defective intra-S checkpoint response in DNA-PKcs-deficient cells. Taken together, these results suggest that DNA-PKcs, in addition to its direct role in DNA damage repair, facilitates ATR-Chk1 signaling pathway in response to replication stress.
Figures

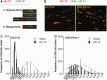
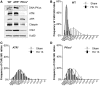
Similar articles
-
Separation of intra-S checkpoint protein contributions to DNA replication fork protection and genomic stability in normal human fibroblasts.Cell Cycle. 2013 Jan 15;12(2):332-45. doi: 10.4161/cc.23177. Epub 2012 Jan 15. Cell Cycle. 2013. PMID: 23255133 Free PMC article.
-
PIDD mediates the association of DNA-PKcs and ATR at stalled replication forks to facilitate the ATR signaling pathway.Nucleic Acids Res. 2018 Feb 28;46(4):1847-1859. doi: 10.1093/nar/gkx1298. Nucleic Acids Res. 2018. PMID: 29309644 Free PMC article.
-
Regulation of ATR-CHK1 signaling by ubiquitination of CLASPIN.Biochem Soc Trans. 2022 Oct 31;50(5):1471-1480. doi: 10.1042/BST20220729. Biochem Soc Trans. 2022. PMID: 36196914 Review.
-
Reconstitution of human claspin-mediated phosphorylation of Chk1 by the ATR (ataxia telangiectasia-mutated and rad3-related) checkpoint kinase.J Biol Chem. 2009 Nov 27;284(48):33107-14. doi: 10.1074/jbc.M109.064485. Epub 2009 Oct 14. J Biol Chem. 2009. PMID: 19828454 Free PMC article.
-
Claspin - checkpoint adaptor and DNA replication factor.FEBS J. 2019 Feb;286(3):441-455. doi: 10.1111/febs.14594. Epub 2018 Jun 29. FEBS J. 2019. PMID: 29931808 Review.
Cited by
-
EGFR Mutations Compromise Hypoxia-Associated Radiation Resistance through Impaired Replication Fork-Associated DNA Damage Repair.Mol Cancer Res. 2017 Nov;15(11):1503-1516. doi: 10.1158/1541-7786.MCR-17-0136. Epub 2017 Aug 11. Mol Cancer Res. 2017. PMID: 28801308 Free PMC article.
-
DNA damage response curtails detrimental replication stress and chromosomal instability induced by the dietary carcinogen PhIP.Nucleic Acids Res. 2016 Dec 1;44(21):10259-10276. doi: 10.1093/nar/gkw791. Epub 2016 Sep 5. Nucleic Acids Res. 2016. PMID: 27599846 Free PMC article.
-
Preserving Genome Integrity During the Early Embryonic DNA Replication Cycles.Genes (Basel). 2019 May 24;10(5):398. doi: 10.3390/genes10050398. Genes (Basel). 2019. PMID: 31137726 Free PMC article. Review.
-
Radiation-dose-dependent functional synergisms between ATM, ATR and DNA-PKcs in checkpoint control and resection in G2-phase.Sci Rep. 2019 Jun 4;9(1):8255. doi: 10.1038/s41598-019-44771-6. Sci Rep. 2019. PMID: 31164689 Free PMC article.
-
USP7 controls Chk1 protein stability by direct deubiquitination.Cell Cycle. 2014;13(24):3921-6. doi: 10.4161/15384101.2014.973324. Cell Cycle. 2014. PMID: 25483066 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

