Connectivity of vertebrate genomes: Paired-related homeobox (Prrx) genes in spotted gar, basal teleosts, and tetrapods
- PMID: 24486528
- PMCID: PMC4032612
- DOI: 10.1016/j.cbpc.2014.01.005
Connectivity of vertebrate genomes: Paired-related homeobox (Prrx) genes in spotted gar, basal teleosts, and tetrapods
Abstract
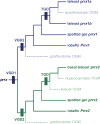
Teleost fish are important models for human biology, health, and disease. Because genome duplication in a teleost ancestor (TGD) impacts the evolution of teleost genome structure and gene repertoires, we must discriminate gene functions that are shared and ancestral from those that are lineage-specific in teleosts or tetrapods to accurately apply inferences from teleost disease models to human health. Generalizations must account both for the TGD and for divergent evolution between teleosts and tetrapods after the likely two rounds of genome duplication shared by all vertebrates. Progress in sequencing techniques provides new opportunities to generate genomic and transcriptomic information from a broad range of phylogenetically informative taxa that facilitate detailed understanding of gene family and gene function evolution. We illustrate here the use of new sequence resources from spotted gar (Lepisosteus oculatus), a rayfin fish that diverged from teleosts before the TGD, as well as RNA-Seq data from gar and multiple teleost lineages to reconstruct the evolution of the Paired-related homeobox (Prrx) transcription factor gene family, which is involved in the development of mesoderm and neural crest-derived mesenchyme. We show that for Prrx genes, the spotted gar genome and gene expression patterns mimic mammals better than teleosts do. Analyses force the seemingly paradoxical conclusion that regulatory mechanisms for the limb expression domains of Prrx genes existed before the evolution of paired appendages. Detailed evolutionary analyses like those reported here are required to identify fish species most similar to the human genome to optimally connect fish models to human gene functions in health and disease.
Keywords: Craniofacial; Fin/limb bud; Genome duplication; Ohnolog; Paired appendages; Prrx1; Prrx2; RNA-Seq.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
A new model army: Emerging fish models to study the genomics of vertebrate Evo-Devo.J Exp Zool B Mol Dev Evol. 2015 Jun;324(4):316-41. doi: 10.1002/jez.b.22589. Epub 2014 Aug 11. J Exp Zool B Mol Dev Evol. 2015. PMID: 25111899 Free PMC article. Review.
-
The Molecular Evolution of Circadian Clock Genes in Spotted Gar (Lepisosteus oculatus).Genes (Basel). 2019 Aug 17;10(8):622. doi: 10.3390/genes10080622. Genes (Basel). 2019. PMID: 31426485 Free PMC article.
-
The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons.Nat Genet. 2016 Apr;48(4):427-37. doi: 10.1038/ng.3526. Epub 2016 Mar 7. Nat Genet. 2016. PMID: 26950095 Free PMC article.
-
Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication.Genetics. 2011 Aug;188(4):799-808. doi: 10.1534/genetics.111.127324. Genetics. 2011. PMID: 21828280 Free PMC article.
-
The Divergent Genomes of Teleosts.Annu Rev Anim Biosci. 2018 Feb 15;6:47-68. doi: 10.1146/annurev-animal-030117-014821. Annu Rev Anim Biosci. 2018. PMID: 29447475 Review.
Cited by
-
Competition between Jagged-Notch and Endothelin1 Signaling Selectively Restricts Cartilage Formation in the Zebrafish Upper Face.PLoS Genet. 2016 Apr 8;12(4):e1005967. doi: 10.1371/journal.pgen.1005967. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27058748 Free PMC article.
-
A new model army: Emerging fish models to study the genomics of vertebrate Evo-Devo.J Exp Zool B Mol Dev Evol. 2015 Jun;324(4):316-41. doi: 10.1002/jez.b.22589. Epub 2014 Aug 11. J Exp Zool B Mol Dev Evol. 2015. PMID: 25111899 Free PMC article. Review.
-
Genome-wide identification of novel ovarian-predominant miRNAs: new insights from the medaka (Oryzias latipes).Sci Rep. 2017 Jan 10;7:40241. doi: 10.1038/srep40241. Sci Rep. 2017. PMID: 28071684 Free PMC article.
-
Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database.BMC Genomics. 2016 May 18;17:368. doi: 10.1186/s12864-016-2709-z. BMC Genomics. 2016. PMID: 27189481 Free PMC article.
-
Na+/Cl- cotransporter 2 is not fish-specific and is widely found in amphibians, non-avian reptiles, and select mammals.Physiol Genomics. 2023 Mar 1;55(3):113-131. doi: 10.1152/physiolgenomics.00143.2022. Epub 2023 Jan 16. Physiol Genomics. 2023. PMID: 36645671 Free PMC article.
References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Amemiya CT, Alfoldi J, Lee AP, Fan S, Philippe H, Maccallum I, Braasch I, Manousaki T, Schneider I, Rohner N, Organ C, Chalopin D, Smith JJ, Robinson M, Dorrington RA, Gerdol M, Aken B, Biscotti MA, Barucca M, Baurain D, Berlin AM, Blatch GL, Buonocore F, Burmester T, Campbell MS, Canapa A, Cannon JP, Christoffels A, De Moro G, Edkins AL, Fan L, Fausto AM, Feiner N, Forconi M, Gamieldien J, Gnerre S, Gnirke A, Goldstone JV, Haerty W, Hahn ME, Hesse U, Hoffmann S, Johnson J, Karchner SI, Kuraku S, Lara M, Levin JZ, Litman GW, Mauceli E, Miyake T, Mueller MG, Nelson DR, Nitsche A, Olmo E, Ota T, Pallavicini A, Panji S, Picone B, Ponting CP, Prohaska SJ, Przybylski D, Saha NR, Ravi V, Ribeiro FJ, Sauka-Spengler T, Scapigliati G, Searle SM, Sharpe T, Simakov O, Stadler PF, Stegeman JJ, Sumiyama K, Tabbaa D, Tafer H, Turner-Maier J, van Heusden P, White S, Williams L, Yandell M, Brinkmann H, Volff JN, Tabin CJ, Shubin N, Schartl M, Jaffe DB, Postlethwait JH, Venkatesh B, Di Palma F, Lander ES, Meyer A, Lindblad-Toh K. The African coelacanth genome provides insights into tetrapod evolution. Nature. 2013;496:311–316. - PMC - PubMed
-
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJ, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. - PubMed
-
- Bergwerff M, Gittenberger-de Groot AC, Wisse LJ, DeRuiter MC, Wessels A, Martin JF, Olson EN, Kern MJ. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000;436:12–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

