Myosin Va mediates Rab8A-regulated GLUT4 vesicle exocytosis in insulin-stimulated muscle cells
- PMID: 24478457
- PMCID: PMC3967978
- DOI: 10.1091/mbc.E13-08-0493
Myosin Va mediates Rab8A-regulated GLUT4 vesicle exocytosis in insulin-stimulated muscle cells
Abstract
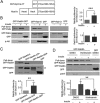
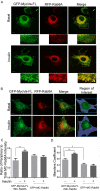
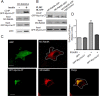
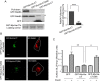
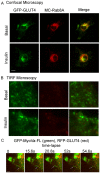
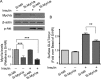
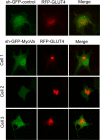
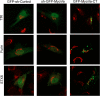
Rab-GTPases are important molecular switches regulating intracellular vesicle traffic, and we recently showed that Rab8A and Rab13 are activated by insulin in muscle to mobilize GLUT4-containing vesicles to the muscle cell surface. Here we show that the unconventional motor protein myosin Va (MyoVa) is an effector of Rab8A in this process. In CHO-IR cell lysates, a glutathione S-transferase chimera of the cargo-binding COOH tail (CT) of MyoVa binds Rab8A and the related Rab10, but not Rab13. Binding to Rab8A is stimulated by insulin in a phosphatidylinositol 3-kinase-dependent manner, whereas Rab10 binding is insulin insensitive. MyoVa-CT preferentially binds GTP-locked Rab8A. Full-length green fluorescent protein (GFP)-MyoVa colocalizes with mCherry-Rab8A in perinuclear small puncta, whereas GFP-MyoVa-CT collapses the GTPase into enlarged perinuclear depots. Further, GFP-MyoVa-CT blocks insulin-stimulated translocation of exofacially myc-tagged GLUT4 to the surface of muscle cells. Mutation of amino acids in MyoVa-CT predicted to bind Rab8A abrogates both interaction with Rab8A (not Rab10) and inhibition of insulin-stimulated GLUT4myc translocation. Of importance, small interfering RNA-mediated MyoVa silencing reduces insulin-stimulated GLUT4myc translocation. Rab8A colocalizes with GLUT4 in perinuclear but not submembrane regions visualized by confocal total internal reflection fluorescence microscopy. Hence insulin signaling to the molecular switch Rab8A connects with the motor protein MyoVa to mobilize GLUT4 vesicles toward the muscle cell plasma membrane.
Figures
Similar articles
-
Rab8A and Rab13 are activated by insulin and regulate GLUT4 translocation in muscle cells.Proc Natl Acad Sci U S A. 2010 Nov 16;107(46):19909-14. doi: 10.1073/pnas.1009523107. Epub 2010 Nov 1. Proc Natl Acad Sci U S A. 2010. PMID: 21041651 Free PMC article.
-
Muscle cells engage Rab8A and myosin Vb in insulin-dependent GLUT4 translocation.Am J Physiol Cell Physiol. 2008 Oct;295(4):C1016-25. doi: 10.1152/ajpcell.00277.2008. Epub 2008 Aug 13. Am J Physiol Cell Physiol. 2008. PMID: 18701652
-
Rab8A regulates insulin-stimulated GLUT4 translocation in C2C12 myoblasts.FEBS Lett. 2017 Feb;591(3):491-499. doi: 10.1002/1873-3468.12555. Epub 2017 Jan 30. FEBS Lett. 2017. PMID: 28079283
-
Update on GLUT4 Vesicle Traffic: A Cornerstone of Insulin Action.Trends Endocrinol Metab. 2017 Aug;28(8):597-611. doi: 10.1016/j.tem.2017.05.002. Epub 2017 Jun 8. Trends Endocrinol Metab. 2017. PMID: 28602209 Review.
-
GLUT4 exocytosis.J Cell Sci. 2011 Dec 15;124(Pt 24):4147-59. doi: 10.1242/jcs.097063. J Cell Sci. 2011. PMID: 22247191 Free PMC article. Review.
Cited by
-
GLUT4 On the move.Biochem J. 2022 Feb 11;479(3):445-462. doi: 10.1042/BCJ20210073. Biochem J. 2022. PMID: 35147164 Free PMC article.
-
Multiple myosin motors interact with sodium/potassium-ATPase alpha 1 subunits.Mol Brain. 2018 Aug 7;11(1):45. doi: 10.1186/s13041-018-0388-1. Mol Brain. 2018. PMID: 30086768 Free PMC article.
-
Thirty sweet years of GLUT4.J Biol Chem. 2019 Jul 26;294(30):11369-11381. doi: 10.1074/jbc.REV119.008351. Epub 2019 Jun 7. J Biol Chem. 2019. PMID: 31175156 Free PMC article. Review.
-
Stimulatory effect on the transport mediated by organic anion transporting polypeptide 2B1.Asian J Pharm Sci. 2020 Mar;15(2):181-191. doi: 10.1016/j.ajps.2019.10.004. Epub 2019 Nov 13. Asian J Pharm Sci. 2020. PMID: 32373198 Free PMC article. Review.
-
Rab35 promotes the recruitment of Rab8, Rab13 and Rab36 to recycling endosomes through MICAL-L1 during neurite outgrowth.Biol Open. 2014 Aug 1;3(9):803-14. doi: 10.1242/bio.20148771. Biol Open. 2014. PMID: 25086062 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

