Radical S-adenosylmethionine enzymes
- PMID: 24476342
- PMCID: PMC4002137
- DOI: 10.1021/cr4004709
Radical S-adenosylmethionine enzymes
Figures
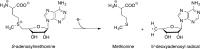
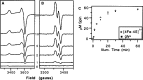
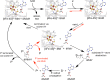
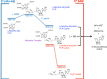
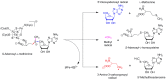
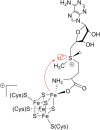
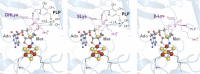
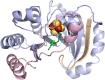

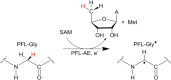
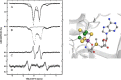
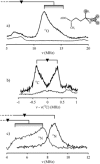
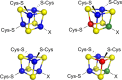
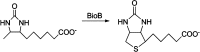
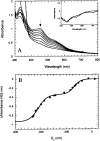
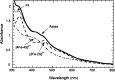
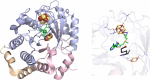

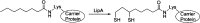
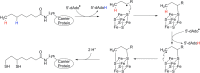
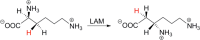

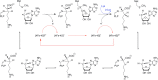

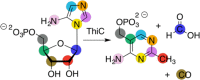
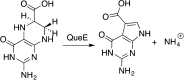
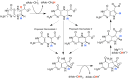
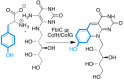
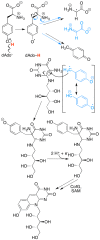
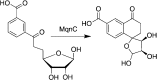
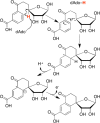

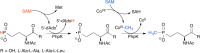
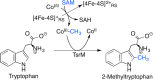
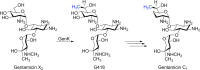
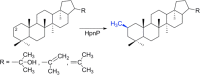




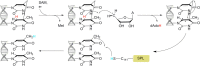
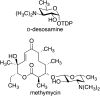
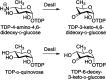
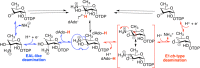
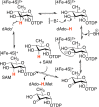
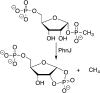
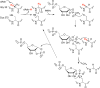
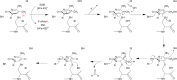
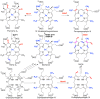
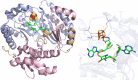
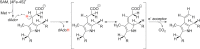
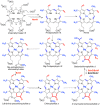
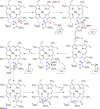
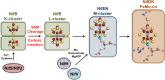
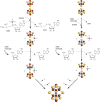
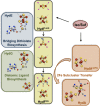
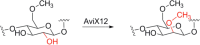

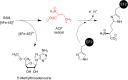
Similar articles
-
Radical SAM enzymes and radical enzymology.Biochim Biophys Acta. 2012 Nov;1824(11):1151-3. doi: 10.1016/j.bbapap.2012.07.006. Epub 2012 Jul 22. Biochim Biophys Acta. 2012. PMID: 22850428 No abstract available.
-
Adenosylmethionine-dependent iron-sulfur enzymes: versatile clusters in a radical new role.J Biol Inorg Chem. 2001 Mar;6(3):209-26. doi: 10.1007/s007750100210. J Biol Inorg Chem. 2001. PMID: 11315557 Review.
-
Pyruvate formate-lyase activating enzyme: elucidation of a novel mechanism for glycyl radical formation.Arch Biochem Biophys. 2005 Jan 1;433(1):288-96. doi: 10.1016/j.abb.2004.09.028. Arch Biochem Biophys. 2005. PMID: 15581584 Review.
-
Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases.Nat Chem Biol. 2013 May;9(5):333-8. doi: 10.1038/nchembio.1229. Epub 2013 Mar 31. Nat Chem Biol. 2013. PMID: 23542644 Free PMC article.
-
The methylthiolation reaction mediated by the Radical-SAM enzymes.Biochim Biophys Acta. 2012 Nov;1824(11):1223-30. doi: 10.1016/j.bbapap.2011.11.007. Epub 2011 Dec 7. Biochim Biophys Acta. 2012. PMID: 22178611 Free PMC article. Review.
Cited by
-
Carbon extension in peptidylnucleoside biosynthesis by radical SAM enzymes.Nat Chem Biol. 2016 Nov;12(11):905-907. doi: 10.1038/nchembio.2187. Epub 2016 Sep 19. Nat Chem Biol. 2016. PMID: 27642865 Free PMC article.
-
S-adenosylmethionine tRNA modification: unexpected/unsuspected implications of former/new players.Int J Biol Sci. 2020 Sep 30;16(15):3018-3027. doi: 10.7150/ijbs.49302. eCollection 2020. Int J Biol Sci. 2020. PMID: 33061813 Free PMC article. Review.
-
Biosynthesis of the catalytic H-cluster of [FeFe] hydrogenase: the roles of the Fe-S maturase proteins HydE, HydF, and HydG.Chem Sci. 2020 Sep 22;11(38):10313-10323. doi: 10.1039/d0sc04216a. Chem Sci. 2020. PMID: 34123177 Free PMC article. Review.
-
Trapping a cross-linked lysine-tryptophan radical in the catalytic cycle of the radical SAM enzyme SuiB.Proc Natl Acad Sci U S A. 2021 May 25;118(21):e2101571118. doi: 10.1073/pnas.2101571118. Proc Natl Acad Sci U S A. 2021. PMID: 34001621 Free PMC article.
-
Radical new paradigm for heme degradation in Escherichia coli O157:H7.Proc Natl Acad Sci U S A. 2016 Oct 25;113(43):12138-12143. doi: 10.1073/pnas.1603209113. Epub 2016 Oct 10. Proc Natl Acad Sci U S A. 2016. PMID: 27791000 Free PMC article.
References
-
- Knappe J.; Schacht J.; Mockel W.; Hopner T.; Vetter H. J.; Edenharder R. Eur. J. Biochem. 1969, 11, 316. - PubMed
-
- Broderick J. B.; Duderstadt R. E.; Fernandez D. C.; Wojtuszewski K.; Henshaw T. F.; Johnson M. K. J. Am. Chem. Soc. 1997, 119, 7396.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

