Activation of ERα signaling differentially modulates IFN-γ induced HLA-class II expression in breast cancer cells
- PMID: 24475282
- PMCID: PMC3903652
- DOI: 10.1371/journal.pone.0087377
Activation of ERα signaling differentially modulates IFN-γ induced HLA-class II expression in breast cancer cells
Abstract
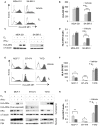
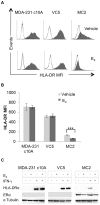
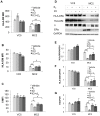
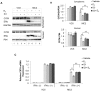
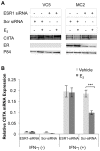
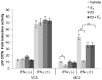
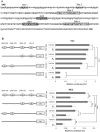
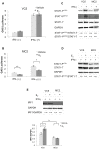
The coordinate regulation of HLA class II (HLA-II) is controlled by the class II transactivator, CIITA, and is crucial for the development of anti-tumor immunity. HLA-II in breast carcinoma is associated with increased IFN-γ levels, reduced expression of the estrogen receptor (ER) and reduced age at diagnosis. Here, we tested the hypothesis that estradiol (E₂) and ERα signaling contribute to the regulation of IFN-γ inducible HLA-II in breast cancer cells. Using a panel of established ER⁻ and ER⁺ breast cancer cell lines, we showed that E₂ attenuated HLA-DR in two ER⁺ lines (MCF-7 and BT-474), but not in T47D, while it augmented expression in ER⁻ lines, SK-BR-3 and MDA-MB-231. To further study the mechanism(s), we used paired transfectants: ERα⁺ MC2 (MDA-MB-231 c10A transfected with the wild type ERα gene) and ERα⁻ VC5 (MDA-MB-231 c10A transfected with the empty vector), treated or not with E₂ and IFN-γ. HLA-II and CIITA were severely reduced in MC2 compared to VC5 and were further exacerbated by E₂ treatment. Reduced expression occurred at the level of the IFN-γ inducible CIITA promoter IV. The anti-estrogen ICI 182,780 and gene silencing with ESR1 siRNA reversed the E2 inhibitory effects, signifying an antagonistic role for activated ERα on CIITA pIV activity. Moreover, STAT1 signaling, necessary for CIITA pIV activation, and selected STAT1 regulated genes were variably downregulated by E₂ in transfected and endogenous ERα positive breast cancer cells, whereas STAT1 signaling was noticeably augmented in ERα⁻ breast cancer cells. Collectively, these results imply immune escape mechanisms in ERα⁺ breast cancer may be facilitated through an ERα suppressive mechanism on IFN-γ signaling.
Conflict of interest statement
Figures

Similar articles
-
Epigenetic inactivation of class II transactivator (CIITA) is associated with the absence of interferon-gamma-induced HLA-DR expression in colorectal and gastric cancer cells.Oncogene. 2004 Nov 25;23(55):8876-86. doi: 10.1038/sj.onc.1208144. Oncogene. 2004. PMID: 15467734
-
Repression of IFN-gamma induction of class II transactivator: a role for PRDM1/Blimp-1 in regulation of cytokine signaling.J Immunol. 2006 Oct 1;177(7):4584-93. doi: 10.4049/jimmunol.177.7.4584. J Immunol. 2006. PMID: 16982896
-
IFN-gamma inducibility of class II transactivator is specifically lacking in human tumour lines: relevance to retinoblastoma protein rescue of IFN-gamma inducibility of the HLA class II genes.Immunol Cell Biol. 1997 Aug;75(4):325-32. doi: 10.1038/icb.1997.50. Immunol Cell Biol. 1997. PMID: 9315472
-
Inactivation of class II transactivator by DNA methylation and histone deacetylation associated with absence of HLA-DR induction by interferon-gamma in haematopoietic tumour cells.Br J Cancer. 2004 Feb 23;90(4):844-52. doi: 10.1038/sj.bjc.6601602. Br J Cancer. 2004. PMID: 14970863 Free PMC article.
-
Dysregulated recruitment of the histone methyltransferase EZH2 to the class II transactivator (CIITA) promoter IV in breast cancer cells.PLoS One. 2012;7(4):e36013. doi: 10.1371/journal.pone.0036013. Epub 2012 Apr 26. PLoS One. 2012. PMID: 22563434 Free PMC article.
Cited by
-
Coordinated inflammation and immune response transcriptional regulation in breast cancer molecular subtypes.Front Immunol. 2024 Jun 25;15:1357726. doi: 10.3389/fimmu.2024.1357726. eCollection 2024. Front Immunol. 2024. PMID: 38983850 Free PMC article.
-
Immunotherapy for Breast Cancer Treatment.Iran Biomed J. 2021 Mar 8;25(3):140-56. doi: 10.29252/ibj.25.3.140. Online ahead of print. Iran Biomed J. 2021. PMID: 33724757 Free PMC article.
-
Therapeutic applications of the selective high affinity ligand drug SH7139 extend beyond non-Hodgkin's lymphoma to many other types of solid cancers.Oncotarget. 2020 Sep 1;11(35):3315-3349. doi: 10.18632/oncotarget.27709. eCollection 2020 Sep 1. Oncotarget. 2020. PMID: 32934776 Free PMC article.
-
Differential expression of the inflammatory ciita gene may be accompanied by altered bone properties in intact sex steroid-deficient female rats.BMC Res Notes. 2023 Dec 19;16(1):372. doi: 10.1186/s13104-023-06543-4. BMC Res Notes. 2023. PMID: 38115045 Free PMC article.
-
A dominant RAD51C pathogenic splicing variant predisposes to breast and ovarian cancer in the Newfoundland population due to founder effect.Mol Genet Genomic Med. 2020 Feb;8(2):e1070. doi: 10.1002/mgg3.1070. Epub 2019 Nov 28. Mol Genet Genomic Med. 2020. PMID: 31782267 Free PMC article.
References
-
- Armstrong TD, Clements VK, Ostrand-Rosenberg S (1998) MHC class II-transfected tumor cells directly present antigen to tumor-specific CD4+ T lymphocytes. J Immunol 160: 661–666. - PubMed
-
- Meazza R, Comes A, Orengo AM, Ferrini S, Accolla RS (2003) Tumor rejection by gene transfer of the MHC class II transactivator in murine mammary adenocarcinoma cells. Eur J Immunol 33: 1183–1192. - PubMed
-
- Accolla RS, Frangione V, De Lerma Barbaro A, Mortara L (2010) New strategies of mammary cancer vaccination. Breast Journal 16: S42–S44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

