Comprehensive transcriptome assembly of Chickpea (Cicer arietinum L.) using sanger and next generation sequencing platforms: development and applications
- PMID: 24465857
- PMCID: PMC3900451
- DOI: 10.1371/journal.pone.0086039
Comprehensive transcriptome assembly of Chickpea (Cicer arietinum L.) using sanger and next generation sequencing platforms: development and applications
Abstract
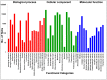
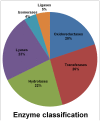
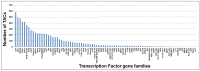
A comprehensive transcriptome assembly of chickpea has been developed using 134.95 million Illumina single-end reads, 7.12 million single-end FLX/454 reads and 139,214 Sanger expressed sequence tags (ESTs) from >17 genotypes. This hybrid transcriptome assembly, referred to as Cicer arietinumTranscriptome Assembly version 2 (CaTA v2, available at http://data.comparative-legumes.org/transcriptomes/cicar/lista_cicar-201201), comprising 46,369 transcript assembly contigs (TACs) has an N50 length of 1,726 bp and a maximum contig size of 15,644 bp. Putative functions were determined for 32,869 (70.8%) of the TACs and gene ontology assignments were determined for 21,471 (46.3%). The new transcriptome assembly was compared with the previously available chickpea transcriptome assemblies as well as to the chickpea genome. Comparative analysis of CaTA v2 against transcriptomes of three legumes - Medicago, soybean and common bean, resulted in 27,771 TACs common to all three legumes indicating strong conservation of genes across legumes. CaTA v2 was also used for identification of simple sequence repeats (SSRs) and intron spanning regions (ISRs) for developing molecular markers. ISRs were identified by aligning TACs to the Medicago genome, and their putative mapping positions at chromosomal level were identified using transcript map of chickpea. Primer pairs were designed for 4,990 ISRs, each representing a single contig for which predicted positions are inferred and distributed across eight linkage groups. A subset of randomly selected ISRs representing all eight chickpea linkage groups were validated on five chickpea genotypes and showed 20% polymorphism with average polymorphic information content (PIC) of 0.27. In summary, the hybrid transcriptome assembly developed and novel markers identified can be used for a variety of applications such as gene discovery, marker-trait association, diversity analysis etc., to advance genetics research and breeding applications in chickpea and other related legumes.
Conflict of interest statement
Figures
Similar articles
-
Large-scale transcriptome analysis in chickpea (Cicer arietinum L.), an orphan legume crop of the semi-arid tropics of Asia and Africa.Plant Biotechnol J. 2011 Oct;9(8):922-31. doi: 10.1111/j.1467-7652.2011.00625.x. Epub 2011 May 25. Plant Biotechnol J. 2011. PMID: 21615673 Free PMC article.
-
Advances in genetics and molecular breeding of three legume crops of semi-arid tropics using next-generation sequencing and high-throughput genotyping technologies.J Biosci. 2012 Nov;37(5):811-20. doi: 10.1007/s12038-012-9228-0. J Biosci. 2012. PMID: 23107917 Review.
-
Development and use of genic molecular markers (GMMs) for construction of a transcript map of chickpea (Cicer arietinum L.).Theor Appl Genet. 2011 May;122(8):1577-89. doi: 10.1007/s00122-011-1556-1. Epub 2011 Mar 8. Theor Appl Genet. 2011. PMID: 21384113 Free PMC article.
-
A comprehensive resource of drought- and salinity- responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.).BMC Genomics. 2009 Nov 15;10:523. doi: 10.1186/1471-2164-10-523. BMC Genomics. 2009. PMID: 19912666 Free PMC article.
-
The developmental dynamics in cool season legumes with focus on chickpea.Plant Mol Biol. 2023 Apr;111(6):473-491. doi: 10.1007/s11103-023-01340-w. Epub 2023 Apr 4. Plant Mol Biol. 2023. PMID: 37016106 Review.
Cited by
-
An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.).Sci Rep. 2015 Aug 11;5:12806. doi: 10.1038/srep12806. Sci Rep. 2015. PMID: 26259924 Free PMC article.
-
Genomics-assisted characterization of a breeding collection of Apios americana, an edible tuberous legume.Sci Rep. 2016 Oct 10;6:34908. doi: 10.1038/srep34908. Sci Rep. 2016. PMID: 27721469 Free PMC article.
-
Contrasting patterns of divergence at the regulatory and sequence level in European Daphnia galeata natural populations.Ecol Evol. 2019 Feb 12;9(5):2487-2504. doi: 10.1002/ece3.4894. eCollection 2019 Mar. Ecol Evol. 2019. PMID: 30891195 Free PMC article.
-
Integrating genomics for chickpea improvement: achievements and opportunities.Theor Appl Genet. 2020 May;133(5):1703-1720. doi: 10.1007/s00122-020-03584-2. Epub 2020 Apr 6. Theor Appl Genet. 2020. PMID: 32253478 Free PMC article. Review.
-
De novo transcriptome assembly of Pueraria montana var. lobata and Neustanthus phaseoloides for the development of eSSR and SNP markers: narrowing the US origin(s) of the invasive kudzu.BMC Genomics. 2018 Jun 5;19(1):439. doi: 10.1186/s12864-018-4798-3. BMC Genomics. 2018. PMID: 29871589 Free PMC article.
References
-
- Brockwell J, Bottomley PJ, Thies JE (1995) Manipulation of rhizobia microflora for improving legume productivity and soil fertility: a critical assessment. In Management of Biological Nitrogen Fixation for the Development of More Productive and Sustainable Agricultural Systems. Springer Netherlands. 143–180.
-
- van Kessel C, Hartley C (2000) Agricultural management of grain legumes: has it led to an increase in nitrogen fixation? Field Crop Res 65(2): 165–181.
-
- Zia-Ul-Haq M, Shahid SA, Ahmed S, Ahmad S, Qayum M, et al. (2012) Anti-platelet activity of methanolic extract of Grewia asiatica L. leaves and Terminalla chebula Retz. fruits. J Med Plants Res 6(10): 2029–2032.
-
- Arumuganathan K, Earle E D (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9(3): 208–218.
-
- Gaur PM, Krishnamurthy L, Kashiwagi J (2008) Improving drought-avoidance root traits in chickpea (Cicer arietinum L.)-current status of research at ICRISAT. Plant Prod Sci 11(1): 3–11.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

