The carboxyl terminus of FANCE recruits FANCD2 to the Fanconi Anemia (FA) E3 ligase complex to promote the FA DNA repair pathway
- PMID: 24451376
- PMCID: PMC3945361
- DOI: 10.1074/jbc.M113.533976
The carboxyl terminus of FANCE recruits FANCD2 to the Fanconi Anemia (FA) E3 ligase complex to promote the FA DNA repair pathway
Abstract
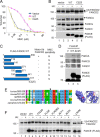
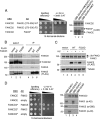
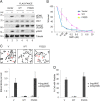
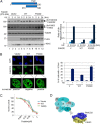
Fanconi anemia (FA) is a genome instability syndrome characterized by bone marrow failure and cellular hypersensitivity to DNA cross-linking agents. In response to DNA damage, the FA pathway is activated through the cooperation of 16 FA proteins. A central player in the pathway is a multisubunit E3 ubiquitin ligase complex or the FA core complex, which monoubiquitinates its substrates FANCD2 and FANCI. FANCE, a subunit of the FA core complex, plays an essential role by promoting the integrity of the complex and by directly recognizing FANCD2. To delineate its role in substrate ubiquitination from the core complex assembly, we analyzed a series of mutations within FANCE. We report that a phenylalanine located at the highly conserved extreme C terminus, referred to as Phe-522, is a critical residue for mediating the monoubiquitination of the FANCD2-FANCI complex. Using the FANCE mutant that specifically disrupts the FANCE-FANCD2 interaction as a tool, we found that the interaction-deficient mutant conferred cellular sensitivity in reconstituted FANCE-deficient cells to a similar degree as FANCE null cells, suggesting the significance of the FANCE-FANCD2 interaction in promoting cisplatin resistance. Intriguingly, ectopic expression of the FANCE C terminus fragment alone in FA normal cells disrupts DNA repair, consolidating the importance of the FANCE-FANCD2 interaction in the DNA cross-link repair.
Keywords: DNA Damage; DNA Repair; E3 Ubiquitin Ligase; Protein Degradation; Protein Dynamics; Protein Folding; Protein Targeting; Protein-protein Interactions; Ubiquitin.
Figures
Similar articles
-
Analysis of a FANCE Splice Isoform in Regard to DNA Repair.J Mol Biol. 2015 Sep 25;427(19):3056-73. doi: 10.1016/j.jmb.2015.08.004. Epub 2015 Aug 12. J Mol Biol. 2015. PMID: 26277624
-
Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway.Mol Cell Biol. 2007 Apr;27(8):3098-108. doi: 10.1128/MCB.02357-06. Epub 2007 Feb 12. Mol Cell Biol. 2007. PMID: 17296736 Free PMC article.
-
Mechanism of Ubiquitination and Deubiquitination in the Fanconi Anemia Pathway.Mol Cell. 2017 Jan 19;65(2):247-259. doi: 10.1016/j.molcel.2016.11.005. Epub 2016 Dec 13. Mol Cell. 2017. PMID: 27986371
-
The ubiquitination machinery of the Fanconi Anemia DNA repair pathway.Prog Biophys Mol Biol. 2021 Aug;163:5-13. doi: 10.1016/j.pbiomolbio.2020.09.009. Epub 2020 Oct 12. Prog Biophys Mol Biol. 2021. PMID: 33058944 Review.
-
Mechanism, specificity, and function of FANCD2-FANCI ubiquitination and deubiquitination.FEBS J. 2022 Aug;289(16):4811-4829. doi: 10.1111/febs.16077. Epub 2021 Jun 29. FEBS J. 2022. PMID: 34137174 Review.
Cited by
-
Damaged mitochondria in Fanconi anemia - an isolated event or a general phenomenon?Oncoscience. 2014 Apr 21;1(4):287-95. doi: 10.18632/oncoscience.29. eCollection 2014. Oncoscience. 2014. PMID: 25594021 Free PMC article.
-
Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.Nucleic Acids Res. 2020 Apr 6;48(6):3328-3342. doi: 10.1093/nar/gkaa062. Nucleic Acids Res. 2020. PMID: 32002546 Free PMC article.
-
FANCJ protein is important for the stability of FANCD2/FANCI proteins and protects them from proteasome and caspase-3 dependent degradation.Oncotarget. 2015 Oct 6;6(30):28816-32. doi: 10.18632/oncotarget.5006. Oncotarget. 2015. PMID: 26336824 Free PMC article.
-
Exploiting the Fanconi Anemia Pathway for Targeted Anti-Cancer Therapy.Mol Cells. 2015 Aug;38(8):669-76. doi: 10.14348/molcells.2015.0175. Epub 2015 Jul 21. Mol Cells. 2015. PMID: 26194820 Free PMC article. Review.
-
Long Noncoding RNA SNHG1 Regulates LMNB2 Expression by Sponging miR-326 and Promotes Cancer Growth in Hepatocellular Carcinoma.Front Oncol. 2021 Nov 30;11:784067. doi: 10.3389/fonc.2021.784067. eCollection 2021. Front Oncol. 2021. PMID: 34917510 Free PMC article.
References
-
- Bogliolo M., Schuster B., Stoepker C., Derkunt B., Su Y., Raams A., Trujillo J. P., Minguillón J., Ramírez M. J., Pujol R., Casado J. A., Baños R., Rio P., Knies K., Zúñiga S., Benítez J., Bueren J. A., Jaspers N. G., Schärer O. D., de Winter J. P., Schindler D., Surrallés J. (2013) Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 92, 800–806 - PMC - PubMed
-
- Osorio A., Bogliolo M., Fernandez V., Barroso A., de la Hoya M., Caldes T., Lasa A., Ramon Y. C. T., Santamarina M., Vega A., Quiles F., Lazaro C., Diez O., Fernandez D., Gonzalez-Sarmiento R., Duran M., Piqueras J. F., Marin M., Pujol R., Surralles J., Benitez J. (2013) Evaluation of Rare Variants in the New Fanconi Anemia Gene ERCC4 (FANCQ) as Familial Breast/Ovarian Cancer Susceptibility Alleles. Hum. Mutat. 34, 1615–1618 - PubMed
-
- Kennedy R. D., D'Andrea A. D. (2005) The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19, 2925–2940 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

