Evidence for a crucial role of a host non-coding RNA in influenza A virus replication
- PMID: 24440876
- PMCID: PMC3929426
- DOI: 10.4161/rna.27504
Evidence for a crucial role of a host non-coding RNA in influenza A virus replication
Abstract
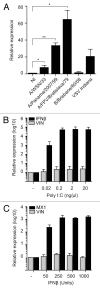
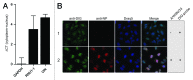
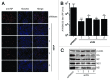
A growing body of evidence suggests the non-protein coding human genome is of vital importance for human cell function. Besides small RNAs, the diverse class of long non-coding RNAs (lncRNAs) recently came into focus. However, their relevance for infection, a major evolutionary driving force, remains elusive. Using two commercially available microarray systems, namely NCode™ and Sureprint™ G3, we identified differential expression of 42 ncRNAs during influenza A virus (IAV) infection in human lung epithelial cells. This included several classes of lncRNAs, including large intergenic ncRNAs (lincRNAs). As analyzed by qRT-PCR, expression of one lincRNA, which we termed virus inducible lincRNA (VIN), is induced by several IAV strains (H1N1, H3N2, H7N7) as well as vesicular stomatitis virus. However, we did not observe an induction of VIN by influenza B virus, treatment with RNA mimics, or IFNβ. Thus, VIN expression seems to be a specific response to certain viral infections. RNA fractionation and RNA-FISH experiments revealed that VIN is localized to the host cell nucleus. Most importantly, we show that abolition of VIN by RNA interference restricts IAV replication and viral protein synthesis, highlighting the relevance of this lincRNA for productive IAV infection. Our observations suggest that viral pathogens interfere with the non-coding portion of the human genome, thereby guaranteeing their successful propagation, and that the expression of VIN correlates with their virulence. Consequently, our study provides a novel approach for understanding virus pathogenesis in greater detail, which will enable future design of new antiviral strategies targeting the host's non-protein coding genome.
Keywords: IAV; VIN; host factor; lincRNA; non-protein coding genome.
Figures


Similar articles
-
Efficient Inhibition of Avian and Seasonal Influenza A Viruses by a Virus-Specific Dicer-Substrate Small Interfering RNA Swarm in Human Monocyte-Derived Macrophages and Dendritic Cells.J Virol. 2019 Feb 5;93(4):e01916-18. doi: 10.1128/JVI.01916-18. Print 2019 Feb 15. J Virol. 2019. PMID: 30463970 Free PMC article.
-
LINC01197 inhibits influenza A virus replication by serving as a PABPC1 decoy.Vet Res. 2024 Sep 27;55(1):121. doi: 10.1186/s13567-024-01379-7. Vet Res. 2024. PMID: 39334466 Free PMC article.
-
β-catenin promotes the type I IFN synthesis and the IFN-dependent signaling response but is suppressed by influenza A virus-induced RIG-I/NF-κB signaling.Cell Commun Signal. 2014 Apr 26;12:29. doi: 10.1186/1478-811X-12-29. Cell Commun Signal. 2014. PMID: 24767605 Free PMC article.
-
Roles of lncRNAs in influenza virus infection.Emerg Microbes Infect. 2020 Dec;9(1):1407-1414. doi: 10.1080/22221751.2020.1778429. Emerg Microbes Infect. 2020. PMID: 32543285 Free PMC article. Review.
-
Involvement of Host Non-Coding RNAs in the Pathogenesis of the Influenza Virus.Int J Mol Sci. 2016 Dec 27;18(1):39. doi: 10.3390/ijms18010039. Int J Mol Sci. 2016. PMID: 28035991 Free PMC article. Review.
Cited by
-
Emerging Role of Interferon-Induced Noncoding RNA in Innate Antiviral Immunity.Viruses. 2022 Nov 23;14(12):2607. doi: 10.3390/v14122607. Viruses. 2022. PMID: 36560611 Free PMC article. Review.
-
Regulation of influenza A virus infection by Lnc-PINK1-2:5.J Cell Mol Med. 2022 Apr;26(8):2285-2298. doi: 10.1111/jcmm.17249. Epub 2022 Feb 24. J Cell Mol Med. 2022. PMID: 35201667 Free PMC article.
-
Analysis of the long noncoding RNA profiles of RD and SH-SY5Y cells infected with coxsackievirus B5, using RNA sequencing.Arch Virol. 2022 Feb;167(2):367-376. doi: 10.1007/s00705-021-05313-6. Epub 2021 Nov 28. Arch Virol. 2022. PMID: 34839420
-
Type I Interferon Regulates the Expression of Long Non-Coding RNAs.Front Immunol. 2014 Nov 6;5:548. doi: 10.3389/fimmu.2014.00548. eCollection 2014. Front Immunol. 2014. PMID: 25414701 Free PMC article.
-
Elucidating the Role of Host Long Non-Coding RNA during Viral Infection: Challenges and Paths Forward.Vaccines (Basel). 2017 Oct 20;5(4):37. doi: 10.3390/vaccines5040037. Vaccines (Basel). 2017. PMID: 29053596 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
